SALT Larval Consensus Summary Statistics
By Dexter Davis - Research Technician for Dr. Shawn Arellano
This Markdown file summarizes the larval data collected with the SyPRID collector on the AUV Sentry during the 2020 EN658 and 2021 TN391 SALT project cruises. This is part of the Seep Animal Larval Transport (SALT) project, NSF Award #1851286.
This markdown file will summarize the data by site as well as by depth classification. Depth classification are as follows: Demersal - 5 meters above bottom, Low Water Column - 80 meters above bottom, Thermocline - depths varied between sites, Mid Water Column - a midway depth between the thermocline and demersal depths.
Data Entry and Subsetting
#install.packages("ggplot2")
library(PNWColors)
library(ggplot2)
library(readxl)
library(knitr)
library(RColorBrewer)
library(gplots)
library(qrcode)
library(vegan)
library(reshape2)
library(ggdendro)
library(plyr)
setwd("C:/Users/davis/OneDrive/Desktop/SPMC Work/Morphotype Consensus")
## Every Dive from Both Cruises
Dive <- read_xlsx("SALT_Larval_Consensus_Final_COPY.xlsx", sheet = "Larval Total Counts")
Dive <- Dive[-c(43),]
Dive <- subset(Dive[-c(7:153)])
## Number of Dives per Site and Depth Classification
Dives <- read_xlsx("SALT_Larval_Consensus_Final_COPY.xlsx", sheet = "Number of Dives")
SiteD <- subset(Dives[-c(1:3)])
colnames(SiteD) <- c("Site","Number of Dives","Average Flow Rate (L/min)")
DepthD <-subset(Dives[-c(4:6)])
DepthD <- na.omit(DepthD)
colnames(DepthD) <- c("Depth Classification","Number of Dives","Average Flow Rate (L/min)")
## Average Volume Collected, Larvae Collected, Larval Density per Site and Depth Classification
Avgs <- read_xlsx("SALT_Larval_Consensus_Final_COPY.xlsx", sheet = "Averages")
SiteAvg <- subset(Avgs[-c(8:15)])
SVol <- subset(SiteAvg[-c(4:7)])
SLar <- subset(SiteAvg[-c(2:3,6:7)])
SDen <- subset(SiteAvg[-c(2:5)])
SDen <- subset(SDen, SDen$Site != "Florida Keys")
DepthAvg <- subset(Avgs[-c(1:8)])
DepthAvg <- na.omit(DepthAvg)
DVol <- subset(DepthAvg[-c(4:7)])
DLar <- subset(DepthAvg[-c(2:3,6:7)])
DDen <- subset(DepthAvg[-c(2:5)])
## Site data untouched - has each site and depth classifications within
Site <- read_xlsx("SALT_Larval_Consensus_Final_COPY.xlsx", sheet = "Larval Density by Site an Depth")
BaltimoreCanyon <- subset(Site,Site$`Sample Site`=="Baltimore Canyon")
BlakeRidge <- subset(Site,Site$`Sample Site`=="Blake Ridge")
BrinePool <- subset(Site,Site$`Sample Site`=="Brine Pool")
BushHill <- subset(Site,Site$`Sample Site`=="Bush Hill")
Chincoteague <- subset(Site,Site$`Sample Site`=="Chincoteague")
FloridaEscarpment <- subset(Site,Site$`Sample Site`=="Florida Escarpment")
GC234 <- subset(Site,Site$`Sample Site`=="Green Canyon 234")
MississippiCanyon <- subset(Site,Site$`Sample Site`=="Mississippi Canyon")
## Larval Type Code Guide
Guide <- read_xlsx("SALT_Larval_Consensus_Final_COPY.xlsx", sheet ="Larval Type Guide")
## Larval Type Presence by Depth
LarvDepth <- read_xlsx("SALT_Larval_Consensus_Final_COPY.xlsx", sheet ="Larval Type by Depth")
LDDem <- subset(LarvDepth,LarvDepth$`Depth Classification`=="Demersal")
LDDem <- subset(LDDem[-c(1)])
LDMWC <- subset(LarvDepth,LarvDepth$`Depth Classification`=="Mid Water Column")
LDMWC <- subset(LDMWC[-c(1)])
LDTherm <- subset(LarvDepth,LarvDepth$`Depth Classification`=="Thermocline")
LDTherm <- subset(LDTherm[-c(1)])Summary of Dives from EN658 and TN391
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
| Cruise | Site | Dive # / Code | Method | Depth (meters, mab= meters above bottom) | Depth Classification |
|---|---|---|---|---|---|
| EN658 | Baltimore Canyon | HN0 | Hand Net | 0 | Surface |
| EN658 | Baltimore Canyon | HN200 | Hand Net | 0-200 | Upper Water Column |
| EN658 | Baltimore Canyon | HN200B | Hand Net | 0-200 | Upper Water Column |
| EN658 | Blake Ridge | S569P | Sentry | 5mab | Demersal |
| EN658 | Blake Ridge | S569S | Sentry | 80mab | Lower Water Column |
| EN658 | Blake Ridge | S570S | Sentry | 5-195mab | Lower Water Column |
| EN658 | Blake Ridge | BR200 | Hand Net | 0-200 | Upper Water Column |
| EN658 | Blake Ridge | BR200B | Hand Net | 0-200 | Upper Water Column |
| EN658 | Brine Pool | S572S | Sentry | 5mab | Demersal |
| EN658 | Brine Pool | S572P | Sentry | 80mab | Mid Water Column |
| EN658 | Brine Pool | BP200 | Hand Net | 0-200 | Upper Water Column |
| EN658 | Brine Pool | BP200B | Hand Net | 0-200 | Upper Water Column |
| EN658 | Bush Hill | BH200B | Hand Net | 0-200 | Upper Water Column |
| EN658 | Bush Hill | BH200C | Hand Net | 0-200 | Upper Water Column |
| EN658 | Florida Keys | FK200 | Hand Net | 0-200 | Upper Water Column |
| EN658 | Florida Keys | FK200B | Hand Net | 0-200 | Upper Water Column |
| EN658 | Mississippi Canyon | MC200 | Hand Net | 0-200 | Upper Water Column |
| EN658 | Mississippi Canyon | MC200B | Hand Net | 0-200 | Upper Water Column |
| TN391 | Baltimore Canyon | S583P | Sentry | 10mab (420) | Demersal |
| TN391 | Baltimore Canyon | S584P | Sentry | 5mab (425) | Demersal |
| TN391 | Baltimore Canyon | S585P | Sentry | 5mab (380) | Demersal |
| TN391 | Baltimore Canyon | S584S | Sentry | 300 | Mid Water Column |
| TN391 | Baltimore Canyon | S583S | Sentry | 200 | Thermocline |
| TN391 | Blake Ridge | S587P | Sentry | 5mab (2160) | Demersal |
| TN391 | Blake Ridge | S588P | Sentry | 5mab (2163) | Demersal |
| TN391 | Blake Ridge | S587S | Sentry | 200 | Thermocline |
| TN391 | Blake Ridge | S588S | Sentry | 200 | Thermocline |
| TN391 | Brine Pool | S592P | Sentry | 5mab (500) | Demersal |
| TN391 | Brine Pool | S593P | Sentry | 5mab | Demersal |
| TN391 | Brine Pool | S592S | Sentry | 300 | Mid Water Column |
| TN391 | Brine Pool | S593S | Sentry | 200 | Thermocline |
| TN391 | Bush Hill | S594P | Sentry | 5mab | Demersal |
| TN391 | Bush Hill | S594S | Sentry | 300 | Mid Water Column |
| TN391 | Chincoteague | S586P | Sentry | 5mab (995) | Demersal |
| TN391 | Florida Escarpment | S589P | Sentry | 5mab (3300) | Demersal |
| TN391 | Florida Escarpment | S590S | Sentry | 1700 | Mid Water Column |
| TN391 | Florida Escarpment | S589S | Sentry | 200 | Thermocline |
| TN391 | Florida Keys | HN200FK | Hand Net | 200 | Surface |
| TN391 | Green Canyon 234 | S591P | Sentry | 5mab (500) | Demersal |
| TN391 | Green Canyon 234 | S591S | Sentry | 300 | Mid Water Column |
| TN391 | Mississippi Canyon | S595P | Sentry | 5mab | Demersal |
| TN391 | Mississippi Canyon | S595S | Sentry | 600 | Mid Water Column |
The rest of this markdown file and data analysis will focus only on the TN391 Sentry dives (this exludes Florida Keys Hand Net) and the EN658 Brine Pool Sentry dives (S572S/S572P). Brine Pool was the only EN658 site that had comparable depths to TN391, and Sentry did not malfunction (EN658 Blake Ridge).
Sentry Performance
Summary Tables of Number of Dives
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
| Site | Number of Dives | Average Flow Rate (L/min) |
|---|---|---|
| Baltimore Canyon | 5 | 11521.929 |
| Blake Ridge | 4 | 10900.721 |
| Brine Pool | 5 | 12619.784 |
| Bush Hill | 2 | 9334.150 |
| Chincoteague | 1 | 11569.161 |
| Florida Escarpment | 3 | 12849.621 |
| Green Canyon 234 | 2 | 11067.122 |
| Mississippi Canyon | 2 | 9261.488 |
Flow Rate Range: 9,261-12,850 Liters/min
| Depth Classification | Number of Dives | Average Flow Rate (L/min) |
|---|---|---|
| Demersal | 13 | 8673.924 |
| Mid Water Column | 6 | 10489.760 |
| Thermocline | 5 | 19611.084 |
Flow Rate Range: 8,674-19,611 Liters/min
Statistics Between Sites and Depth Classifications
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
Sites
Volume Sampled
#SVol
my_title <- "Average Volume of Water Collected by SyPRID by Site"
ggplot(SVol, aes(x=SVol$Site, y=SVol$`Avg Volume Sampled`))+geom_bar(fill="deepskyblue3",stat="identity",position=position_dodge(),width = 0.9)+labs(title =my_title, x="Site", y="Average Volume Collected (L)")+theme(plot.title = element_text(hjust=0.5), axis.text.x=element_text(angle = 45, vjust = 1, hjust=1))+geom_errorbar(aes(ymin=SVol$`Avg Volume Sampled`- SVol$`Volume STDev...3`, ymax=SVol$`Avg Volume Sampled`+ SVol$`Volume STDev...3`), width=.2,position=position_dodge(.9))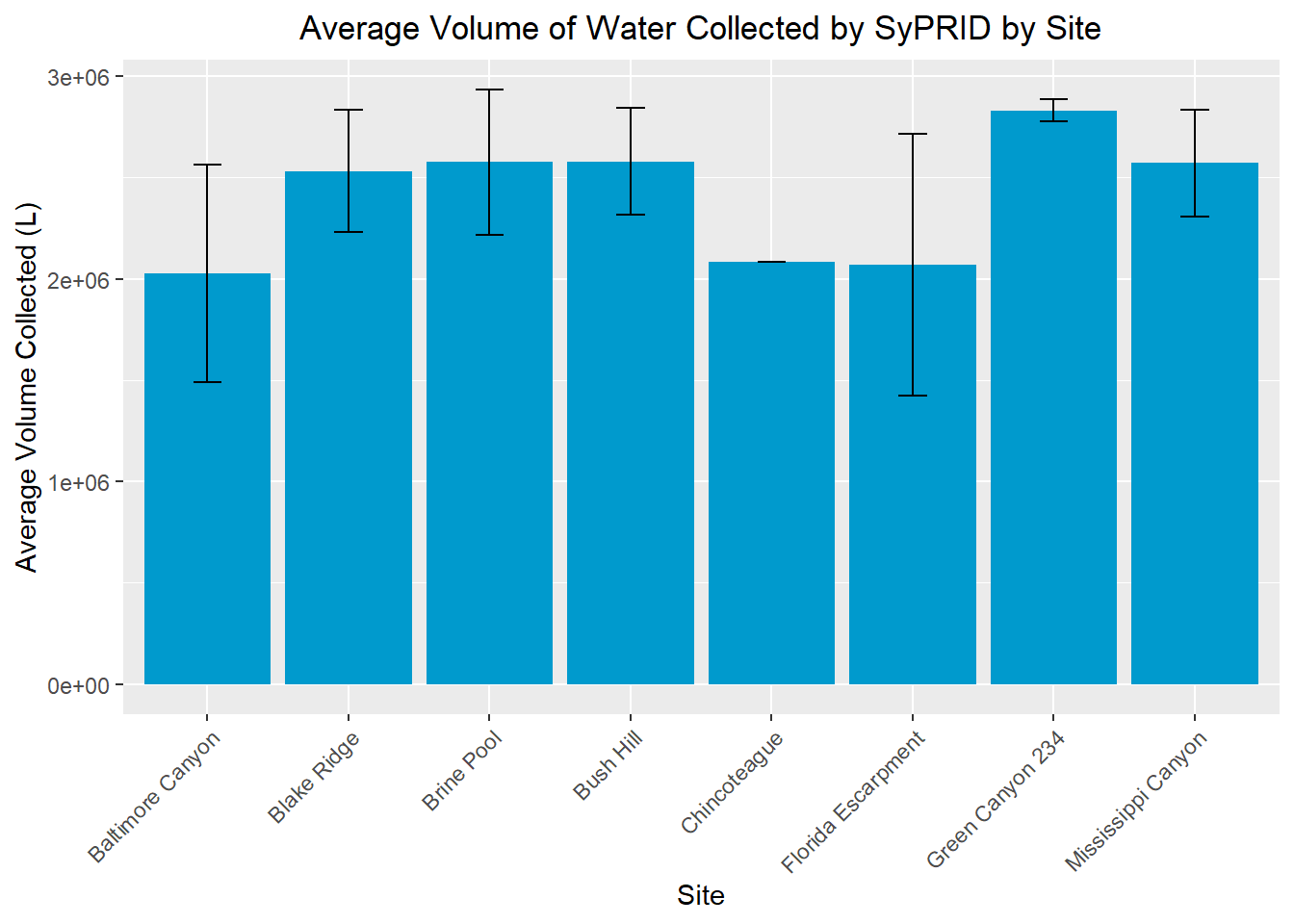
Larvae Collected
#SLAr
my_title <- "Average Number of Larvae Collected by SyPRID by Site"
ggplot(SLar, aes(x=SLar$Site, y=SLar$`Avg #Larva Collected`))+geom_bar(fill="palevioletred3",stat="identity",position=position_dodge(),width = 0.9)+labs(title =my_title, x="Site", y="Average # Larvae Collected")+theme(plot.title = element_text(hjust=0.5), axis.text.x=element_text(angle = 45, vjust = 1, hjust=1))+geom_errorbar(aes(ymin=SLar$`Avg #Larva Collected`- SLar$`Larvae STDev...5`, ymax=SLar$`Avg #Larva Collected`+SLar$`Larvae STDev...5`), width=.2,position=position_dodge(.9))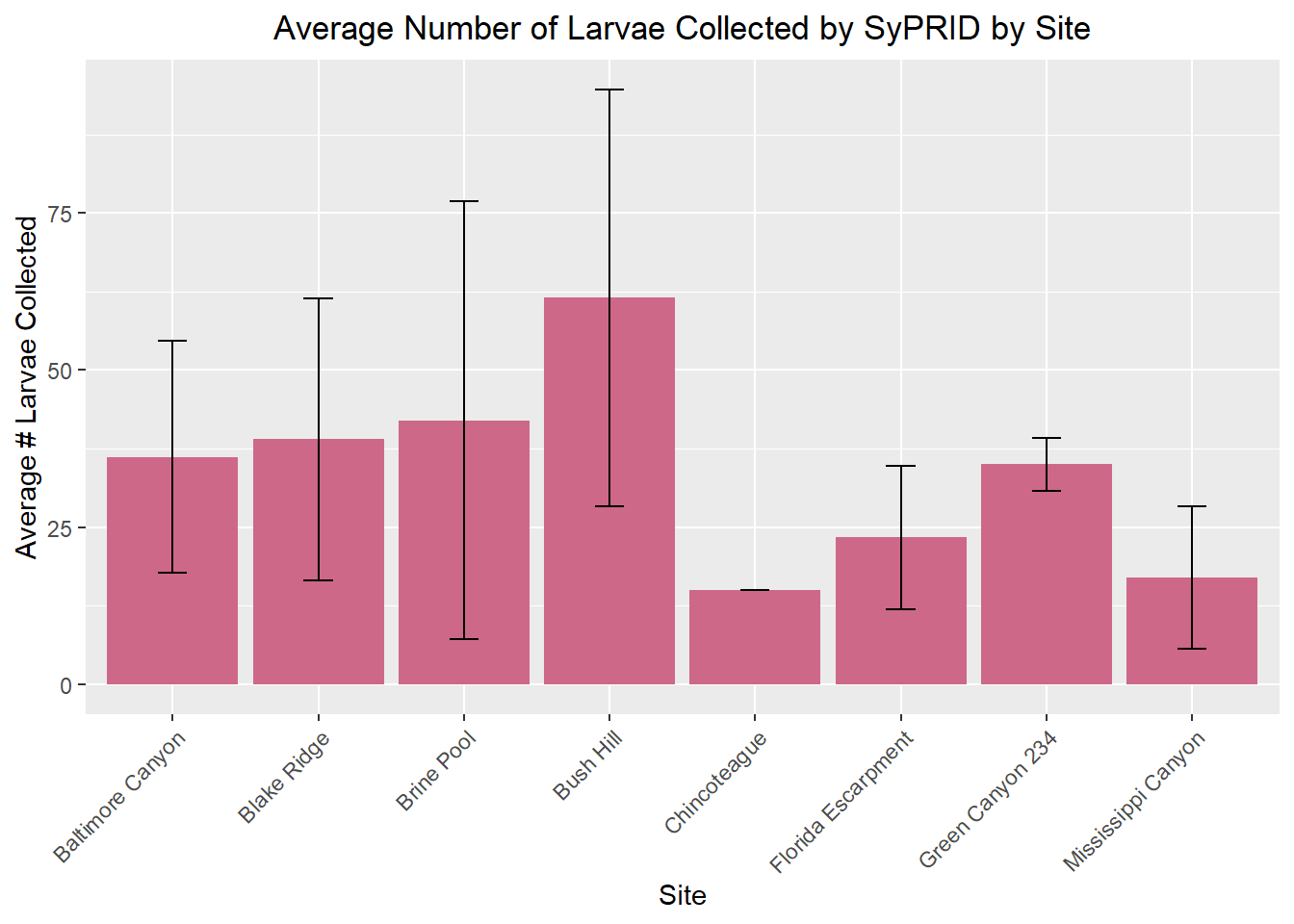
Larval Density
#SDen
SDen$Site <- factor(SDen$Site,levels=c("Baltimore Canyon","Chincoteague","Blake Ridge","Florida Escarpment","Mississippi Canyon","Green Canyon 234","Bush Hill","Brine Pool"))
my_title <- "Average Larval Density Collected by SyPRID by Site"
ggplot(SDen, aes(x=SDen$Site, y=SDen$`Avg Larval Density`,fill=SDen$Site))+geom_bar(stat="identity",position=position_dodge(),width = 0.9)+labs(title =my_title, x="Site", y="Average Larval Density (#/L)")+theme(plot.title = element_text(hjust=0.5), axis.text.x=element_text(angle = 45, vjust = 1, hjust=1))+geom_errorbar(aes(ymin=SDen$`Avg Larval Density`- SDen$`Density STDev...7`, ymax=SDen$`Avg Larval Density`+SDen$`Density STDev...7`), width=.2,position=position_dodge(.9))+scale_fill_viridis_d()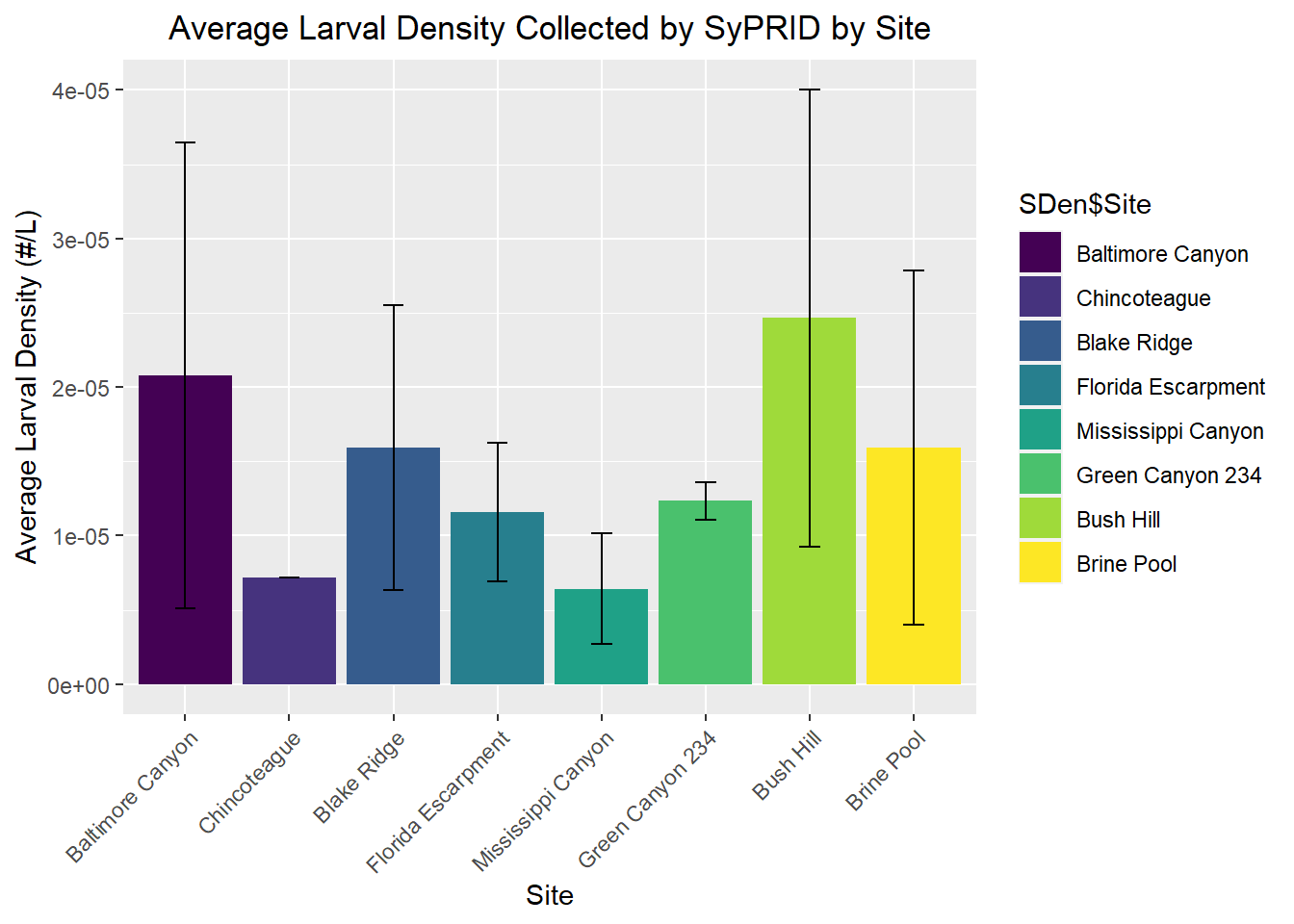
Depth Classifications
Volume Sampled
#DVol
my_title <- "Average Volume of Water Collected by SyPRID by Depth Classification"
ggplot(DVol, aes(x=DVol$`Depth Classification`, y=DVol$`Avg. Volume Sampled (L)`))+geom_bar(fill="deepskyblue3",stat="identity",position=position_dodge(),width = 0.9)+labs(title =my_title, x="Depth", y="Average Volume Collected (L)")+theme(plot.title = element_text(hjust=0.5), axis.text.x=element_text(angle = 45, vjust = 1, hjust=1))+geom_errorbar(aes(ymin=DVol$`Avg. Volume Sampled (L)`- DVol$`Volume STDev...11`, ymax=DVol$`Avg. Volume Sampled (L)`+ DVol$`Volume STDev...11`), width=.2,position=position_dodge(.9))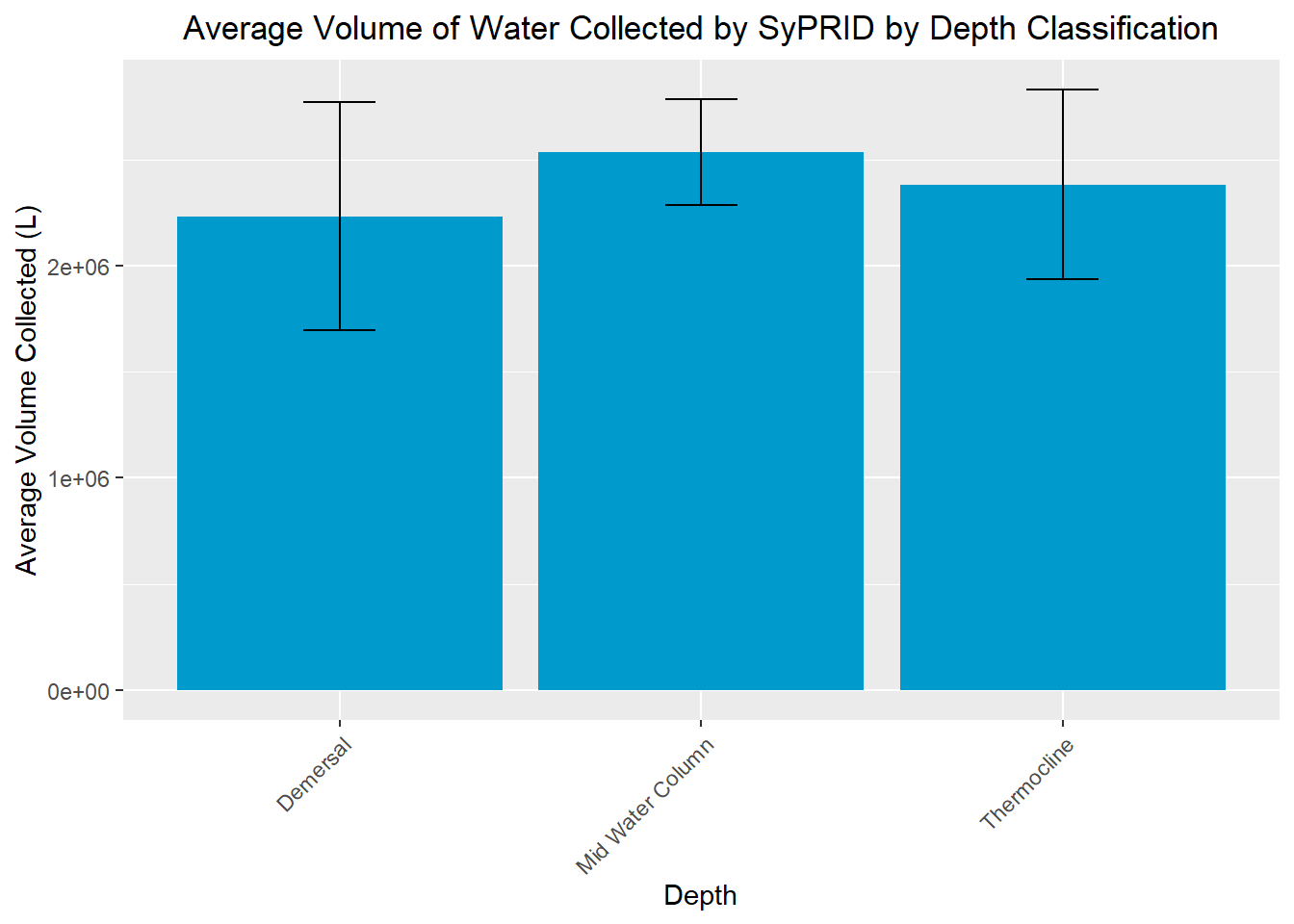
Larvae Collected
#DLar
my_title <- "Average Number of Larvae Collected by SyPRID by Depth Classification"
ggplot(DLar, aes(x=DLar$`Depth Classification`, y=DLar$`Avg. Larvae Collected`))+geom_bar(fill="palevioletred3",stat="identity",position=position_dodge(),width = 0.9)+labs(title =my_title, x="Depth", y="Average # Larvae Collected")+theme(plot.title = element_text(hjust=0.5), axis.text.x=element_text(angle = 45, vjust = 1, hjust=1))+geom_errorbar(aes(ymin=DLar$`Avg. Larvae Collected`- DLar$`Larvae STDev...13`, ymax=DLar$`Avg. Larvae Collected`+DLar$`Larvae STDev...13`), width=.2,position=position_dodge(.9))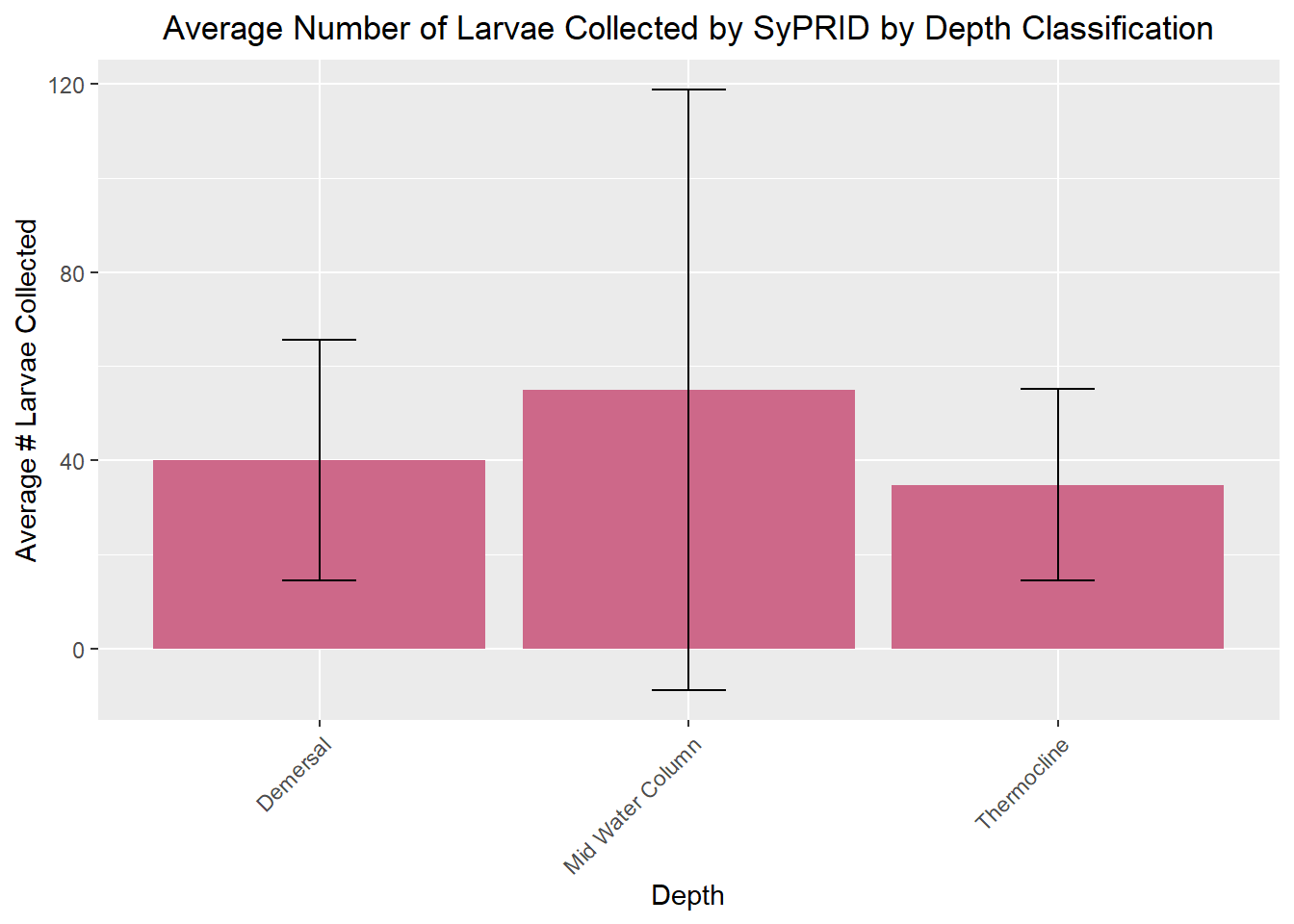
Larval Density
#DDen
my_title <- "Average Larval Density Collected by SyPRID by Depth Classification"
ggplot(DDen, aes(x=DDen$`Depth Classification`, y=DDen$`Avg. Larval Density (#/L)`))+geom_bar(fill=c("darkslategray3","blueviolet","dodgerblue4"),stat="identity",position=position_dodge(),width = 0.9)+labs(title =my_title, x="Site", y="Average Larval Density (#/L)")+theme(plot.title = element_text(hjust=0.5), axis.text.x=element_text(angle = 45, vjust = 1, hjust=1))+geom_errorbar(aes(ymin=DDen$`Avg. Larval Density (#/L)`- DDen$`Density STDev...15`, ymax=DDen$`Avg. Larval Density (#/L)`+DDen$`Density STDev...15`), width=.2,position=position_dodge(.9))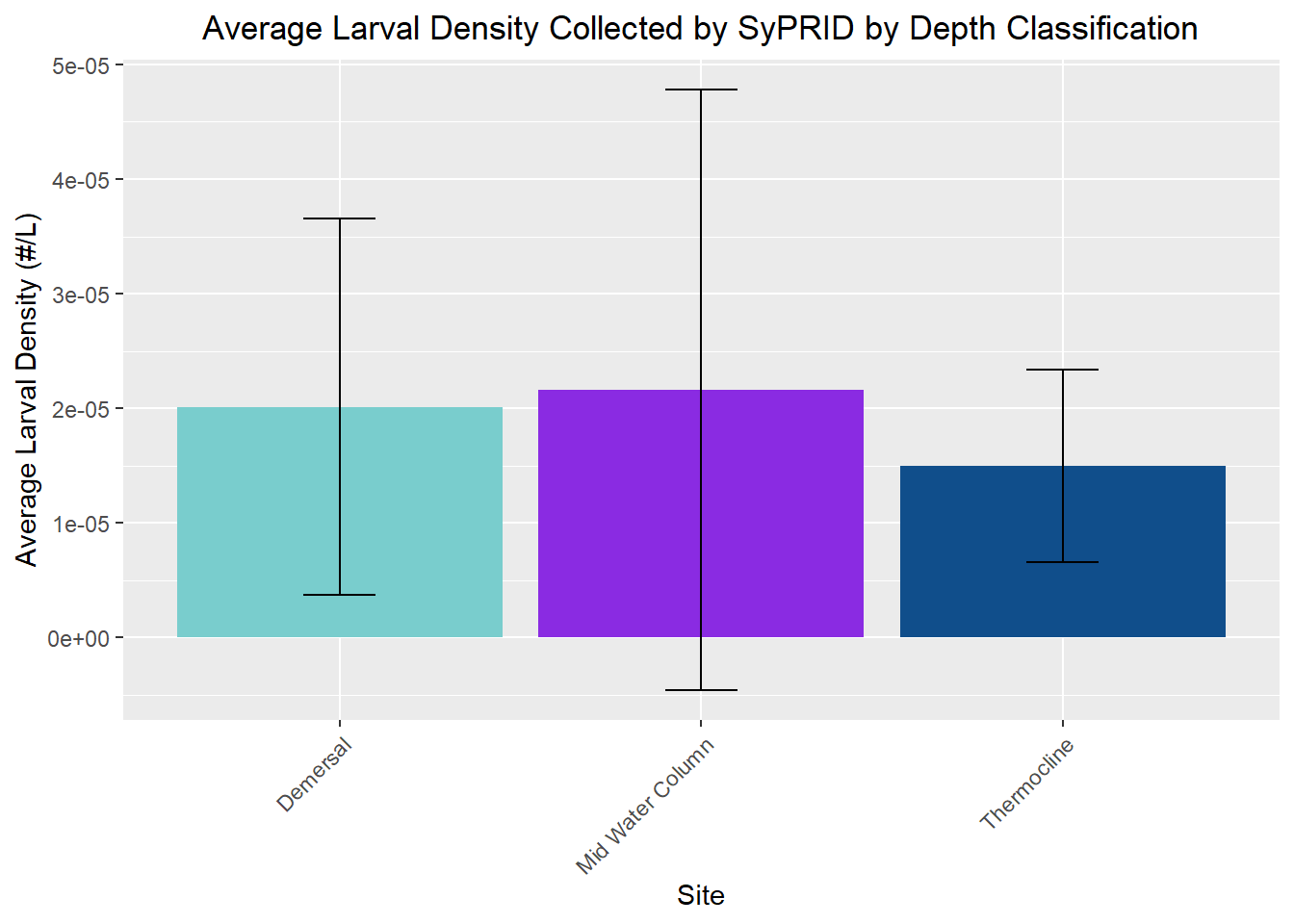
Statistics Within Individual Sites
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
For sites that had multiple dives at given depths (ex. 3 Demersal depths at Brine Pool), the values were averaged resulting in one bar per depth per site.
#Let's make some fuctions!
wrapper <- function(x, ...)
{
paste(strwrap(x, ...), collapse = "\n")
}
#Volume Sampled
Vol <- function(site,title){
ggplot(site, aes(x=`Depth Classification`,y=`Volume Sampled (L)`))+geom_bar(stat="identity", fill='deepskyblue3')+theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust=1))+labs(title=paste0('Volume Sampled with SyPRID by Depth Classificiation at ',title),x="Depth Classificiation", y="Volume Sampled (L)")+theme(plot.title = element_text(hjust = 0.5))
}
#Larvae Collected
Lar <- function(site,title){
ggplot(site, aes(x=`Depth Classification`,y=`Total Larva Collected`))+geom_bar(stat="identity", fill='palevioletred3')+theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust=1))+labs(title=paste0('Larvae Collected with SyPRID by Depth Classification at ',title),x="Depth Classificiation", y="Number of Larvae")+theme(plot.title = element_text(hjust = 0.5))
}
#Larval Density
Den <- function(site,title){
my_title <- paste0('Larval Density collected with SyPRID by Depth Classification at ',title)
ggplot(site, aes(x=`Depth Classification`,y=`Larval Density (#/L)`))+geom_bar(stat="identity", fill='salmon2')+theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust=1))+labs(title=my_title ,x="Depth Classificiation", y="Larval Density (#/L)")+theme(plot.title = element_text(hjust = 0.5))+ggtitle(wrapper(my_title, width = 70))
} Baltimore Canyon
Volume Sampled
Vol(BaltimoreCanyon,"Baltimore Canyon")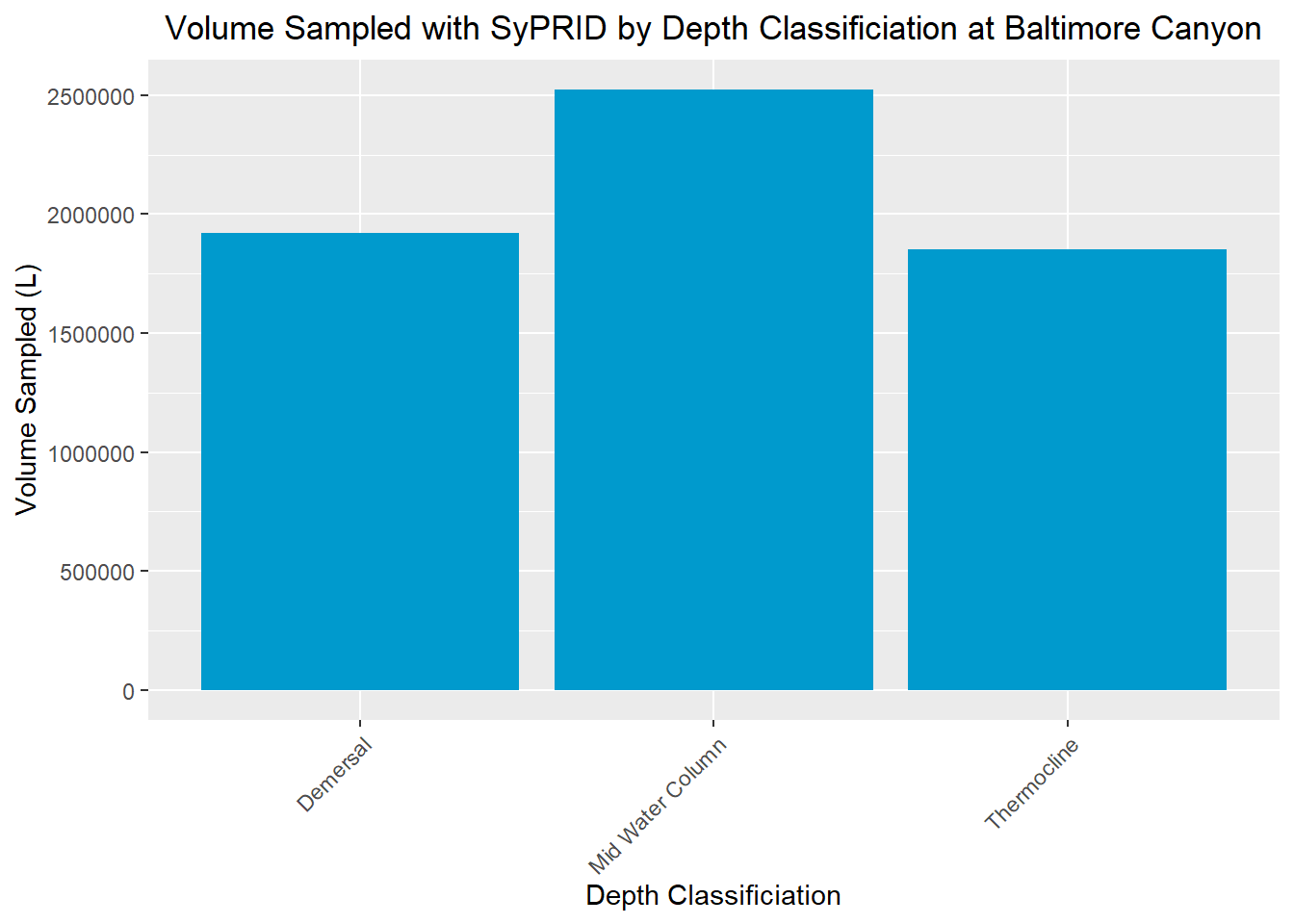
Larvae Collected
Lar(BaltimoreCanyon,"Baltimore Canyon")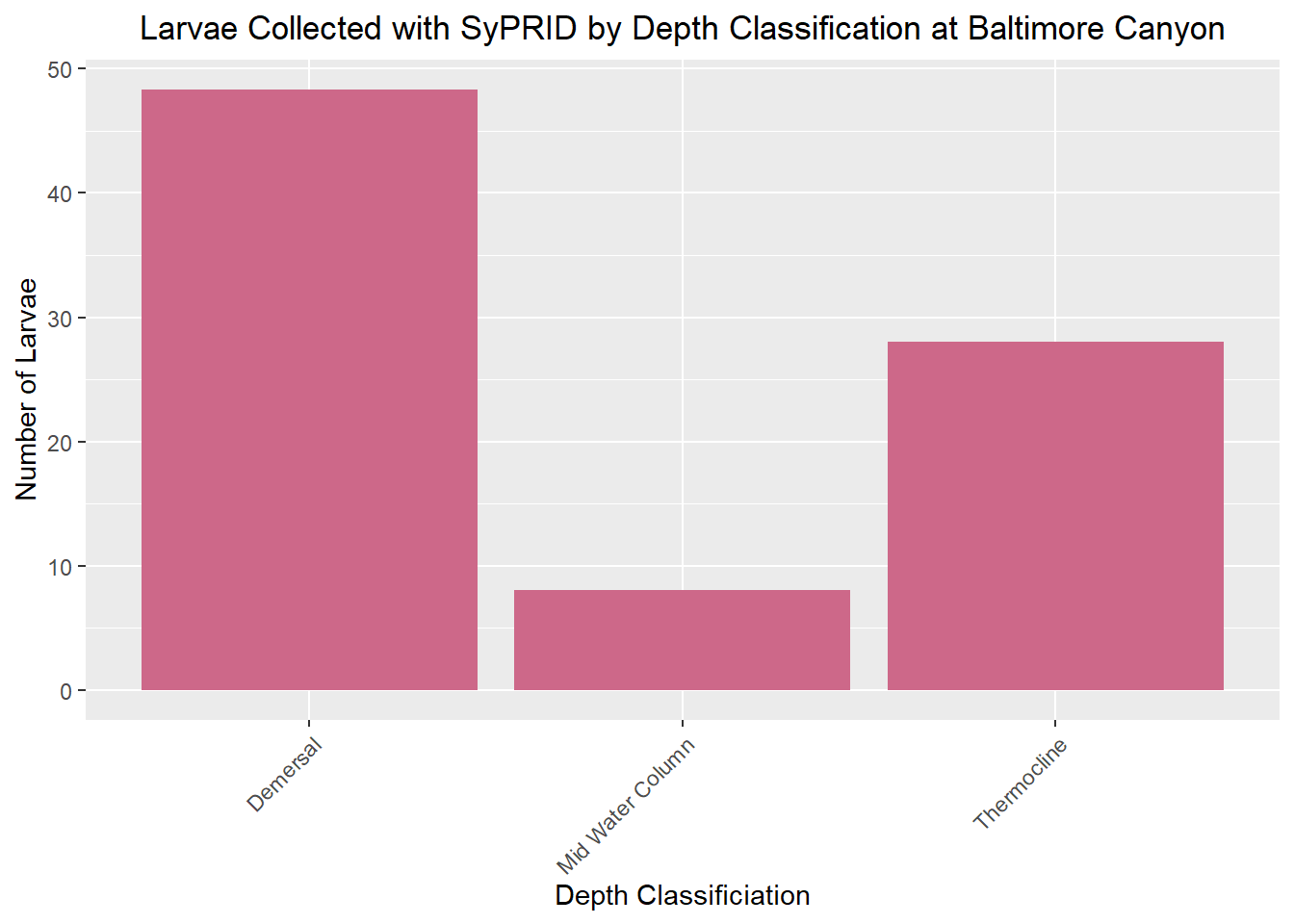
Larval Density
Den(BaltimoreCanyon,"Baltimore Canyon")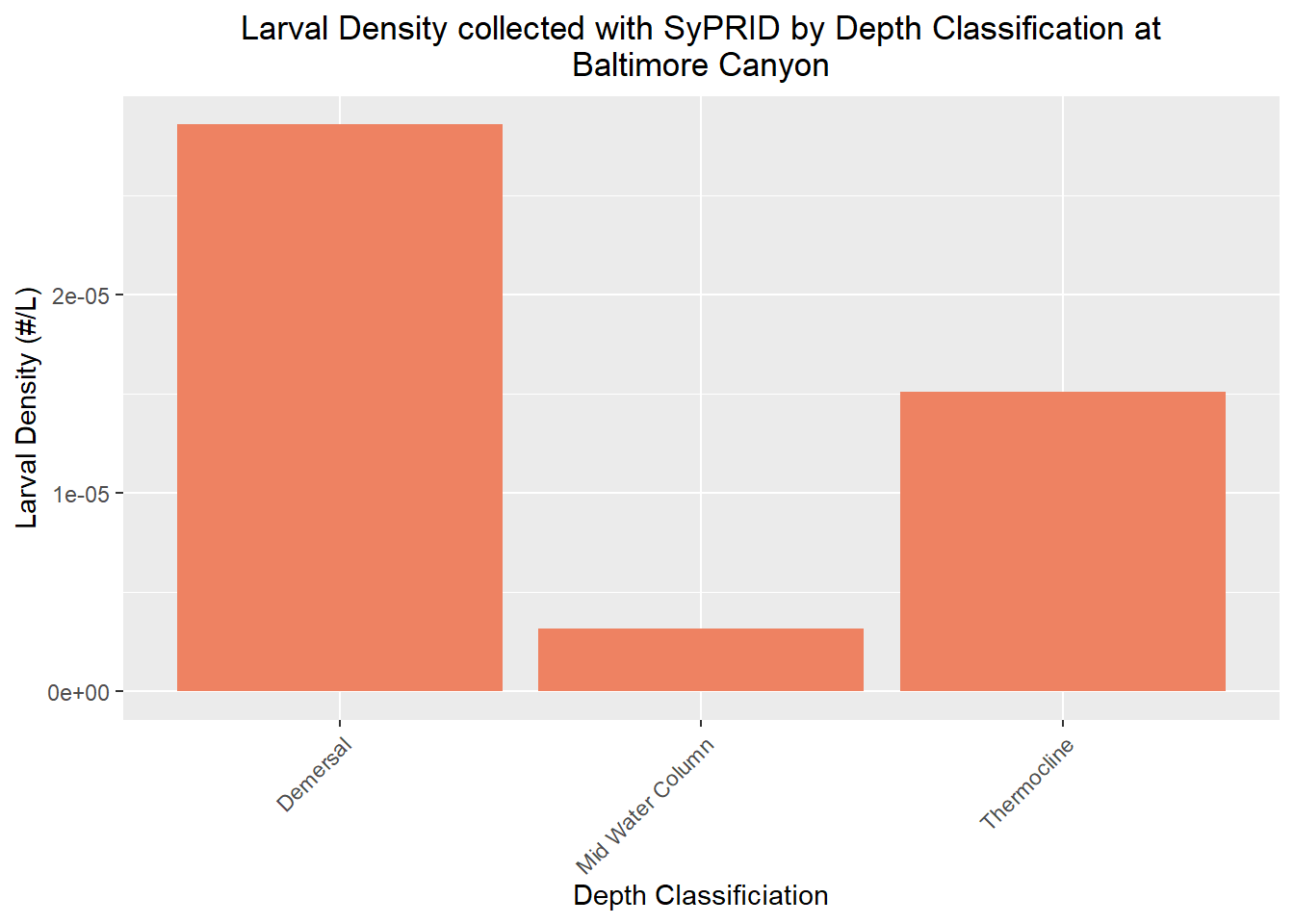
Blake Ridge
Volume Sampled
Vol(BlakeRidge, "Blake Ridge")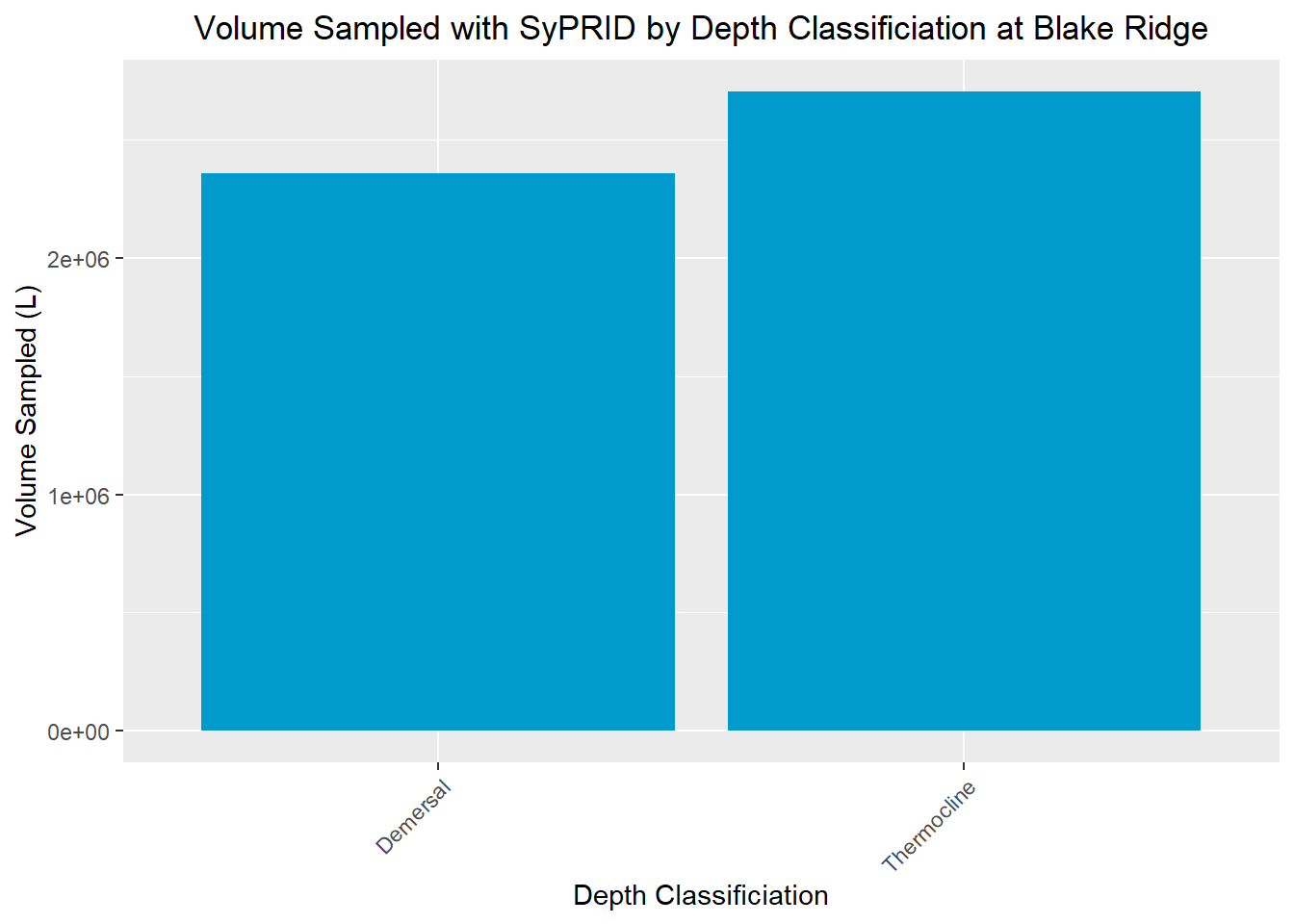
Larvae Collected
Lar(BlakeRidge, "Blake Ridge")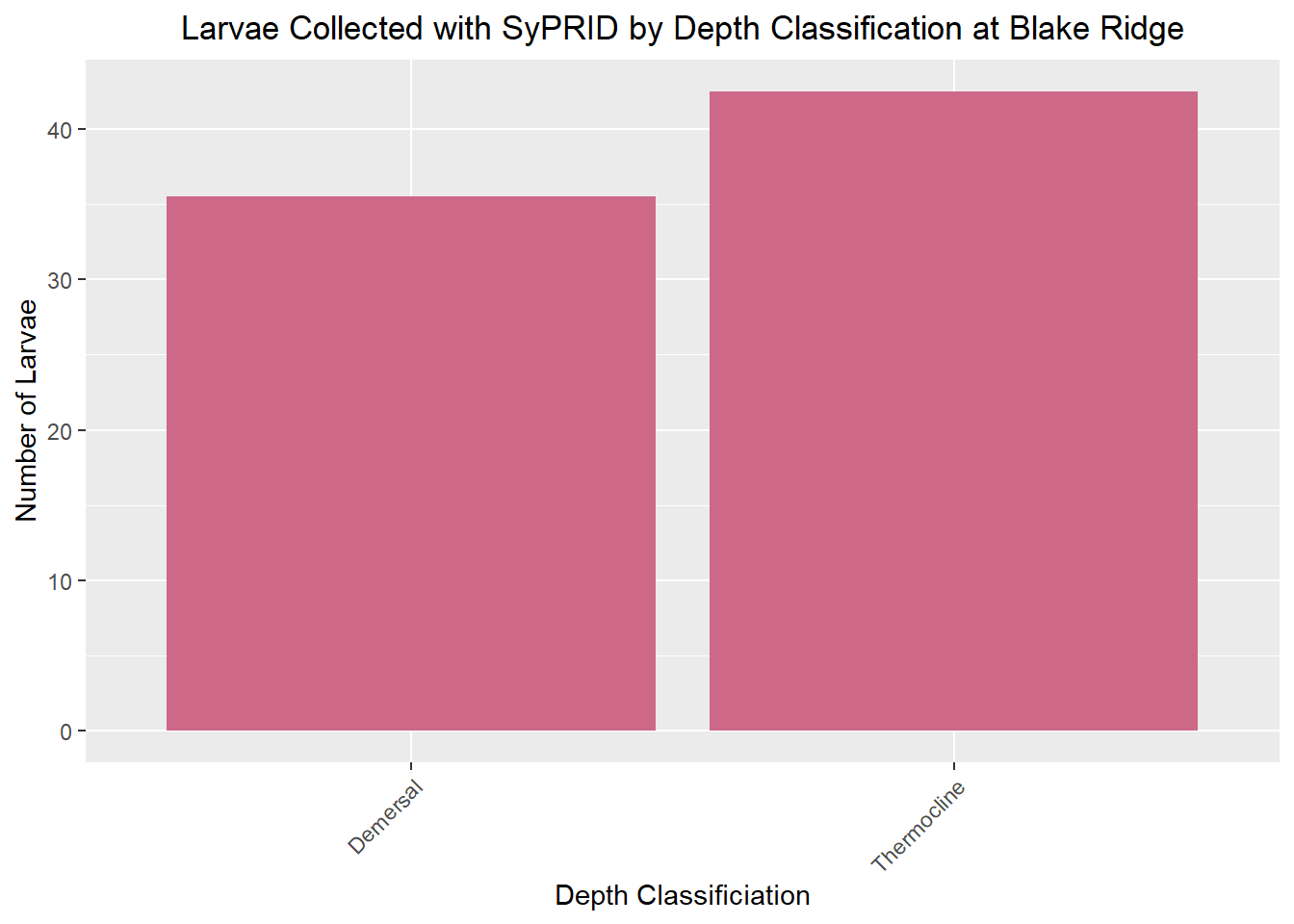
Larval Density
Den(BlakeRidge, "Blake Ridge")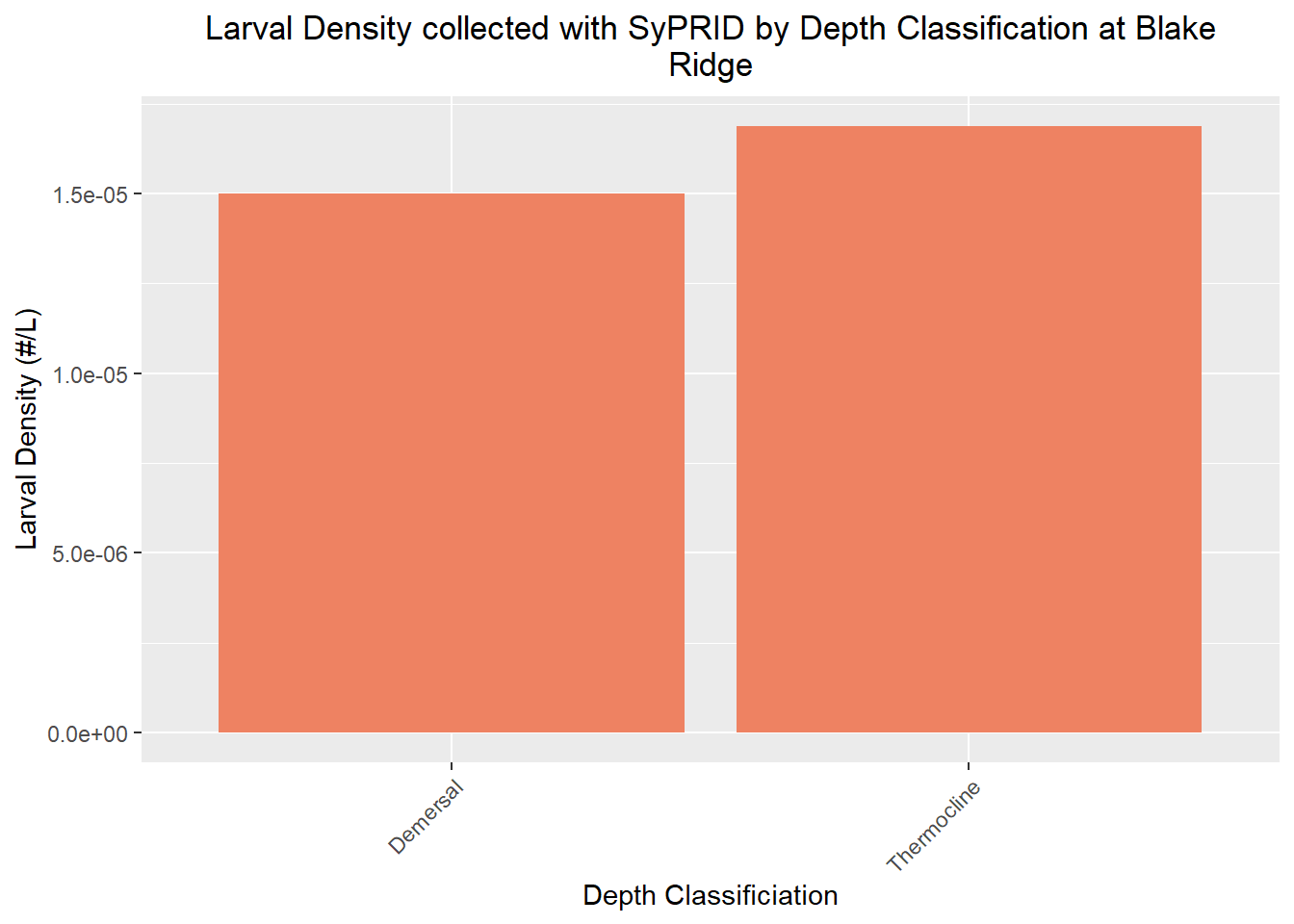
Brine Pool
Volume Sampled
Vol(BrinePool, "Brine Pool")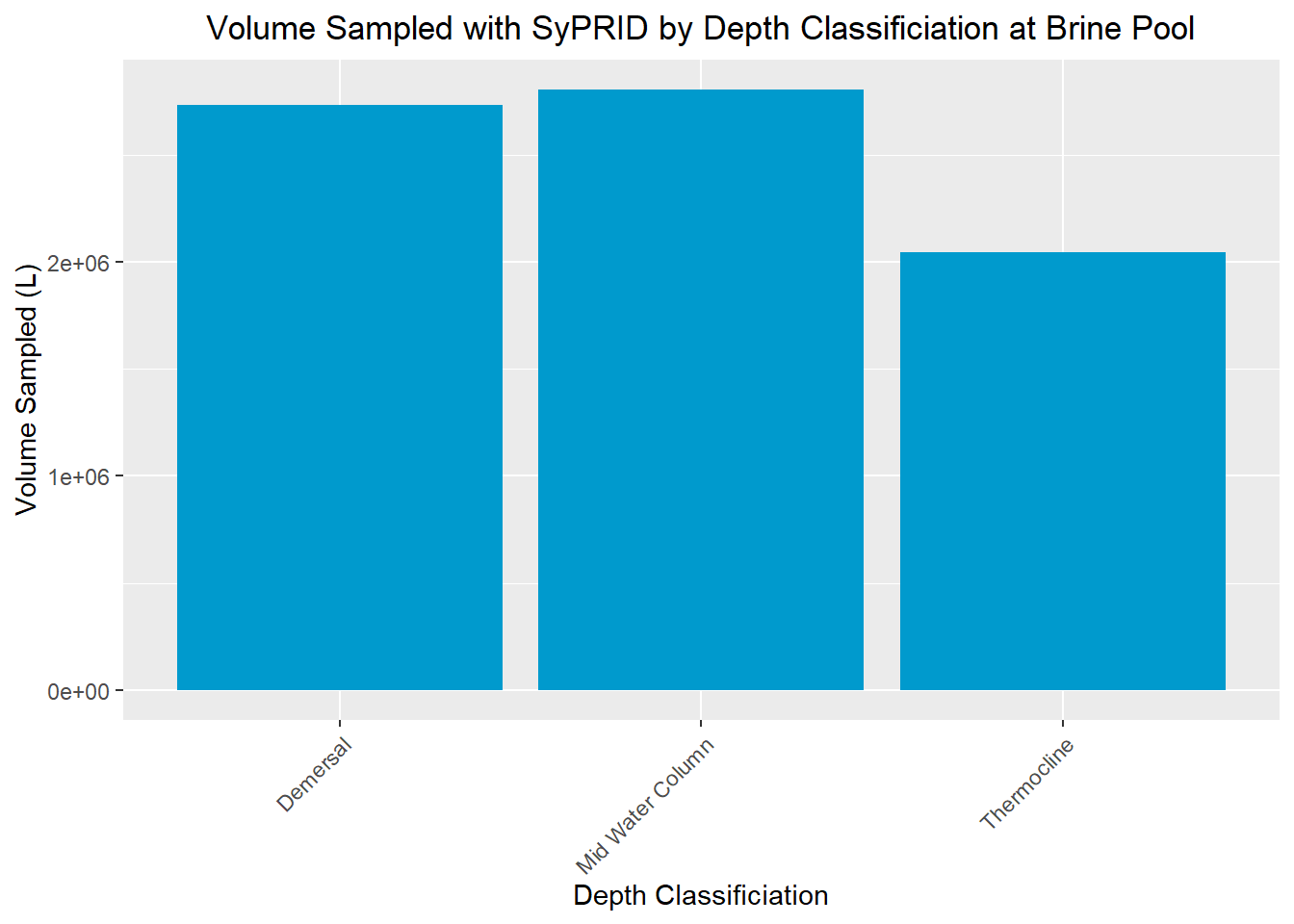
Larvae Collected
Lar(BrinePool, "Brine Pool")
Larval Density
Den(BrinePool, "Brine Pool")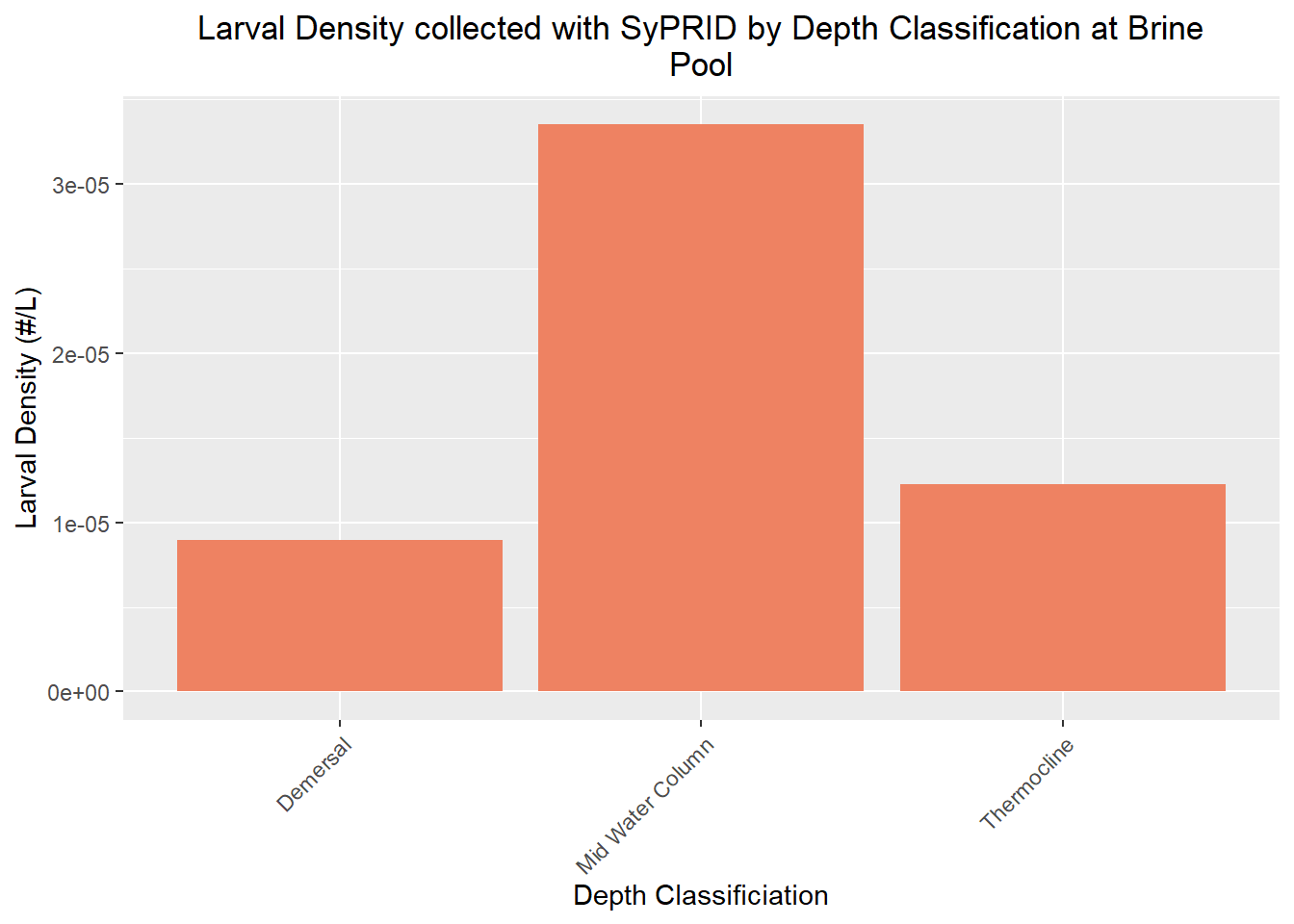
Bush Hill
Volume Sampled
Vol(BushHill, "Bush Hill")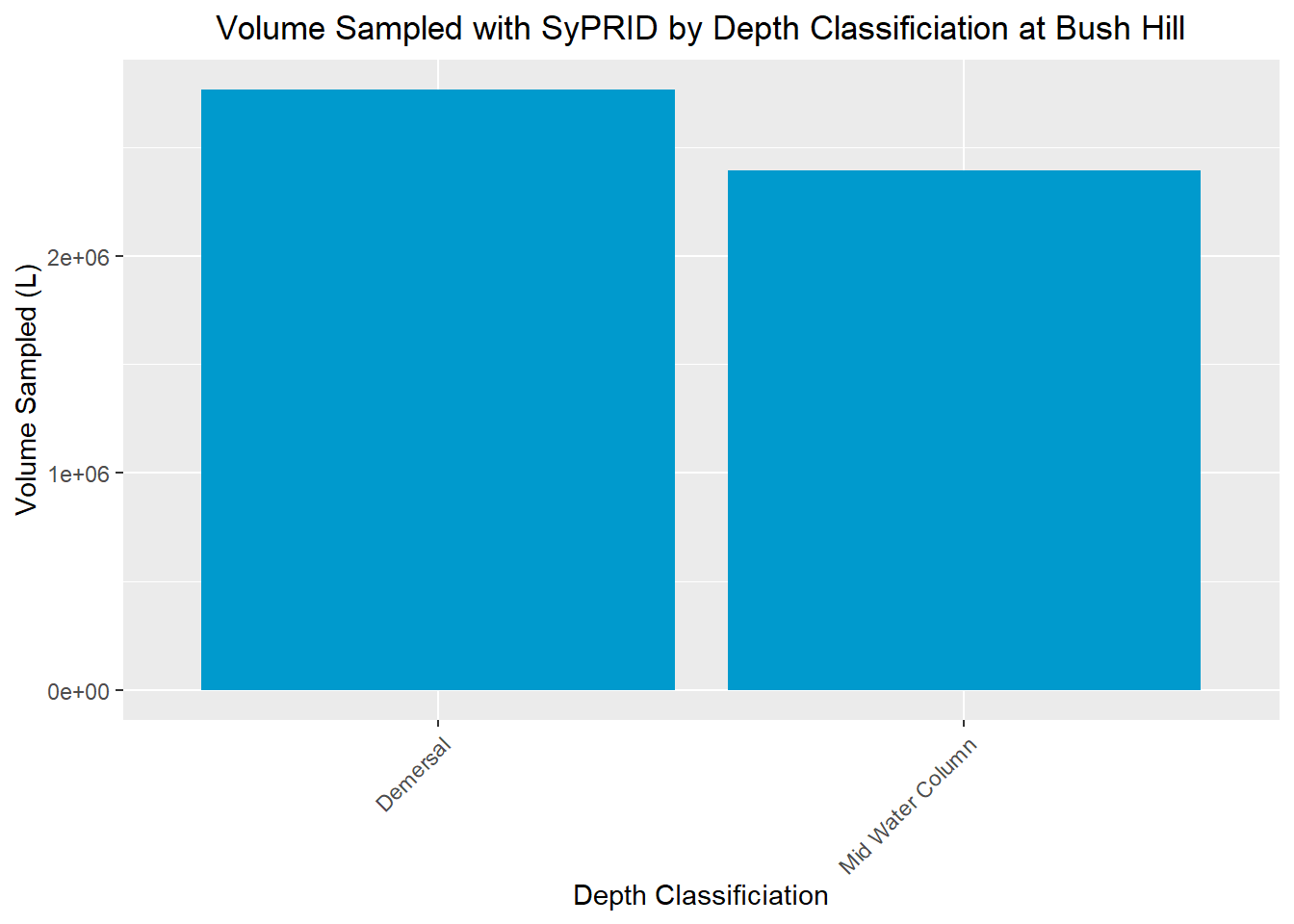
Larvae Collected
Lar(BushHill, "Bush Hill")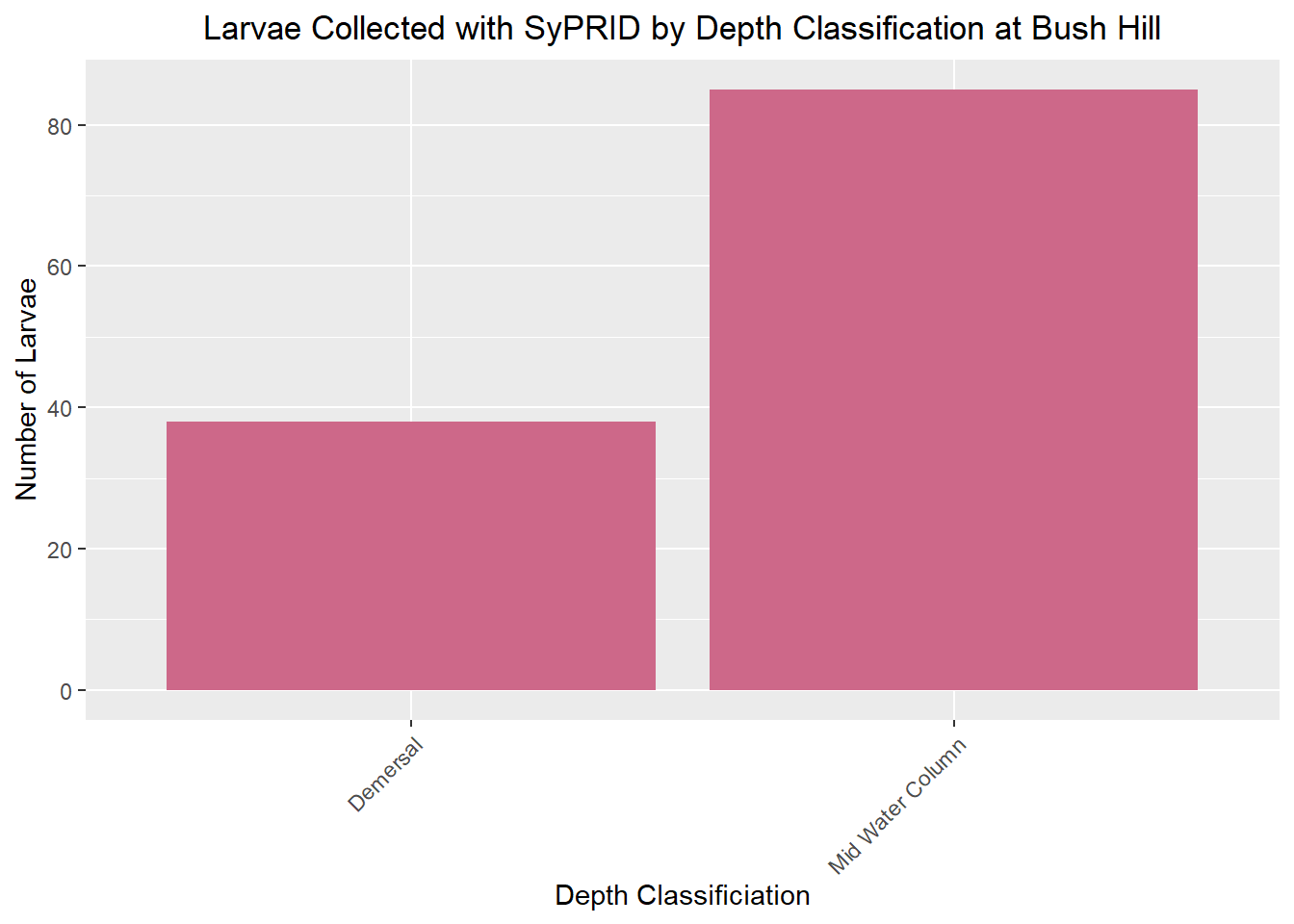
Larval Density
Den(BushHill, "Bush Hill")
Chincoteague
Volume Sampled
Vol(Chincoteague, "Chincoteague")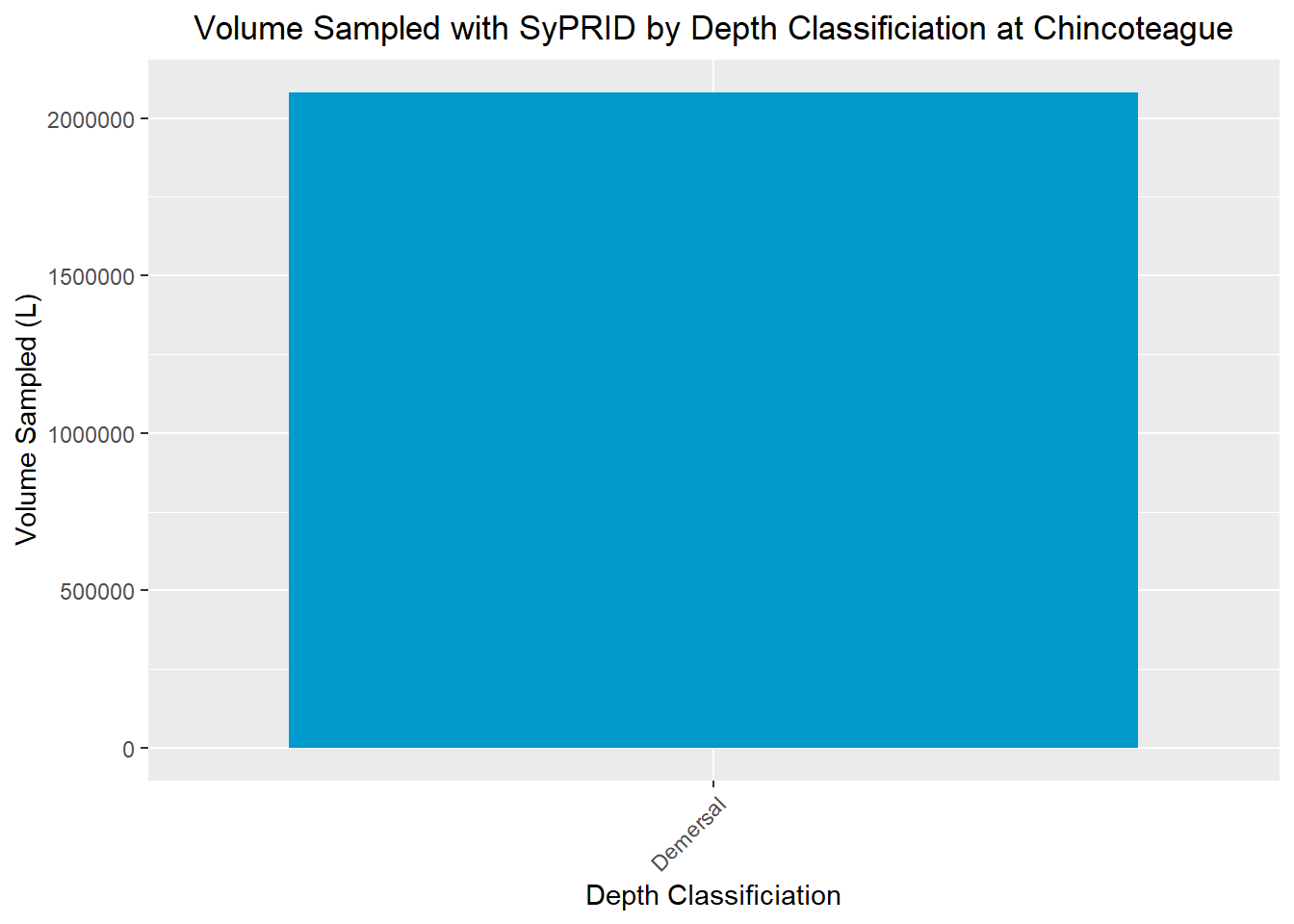
Larvae Collected
Lar(Chincoteague, "Chincoteague")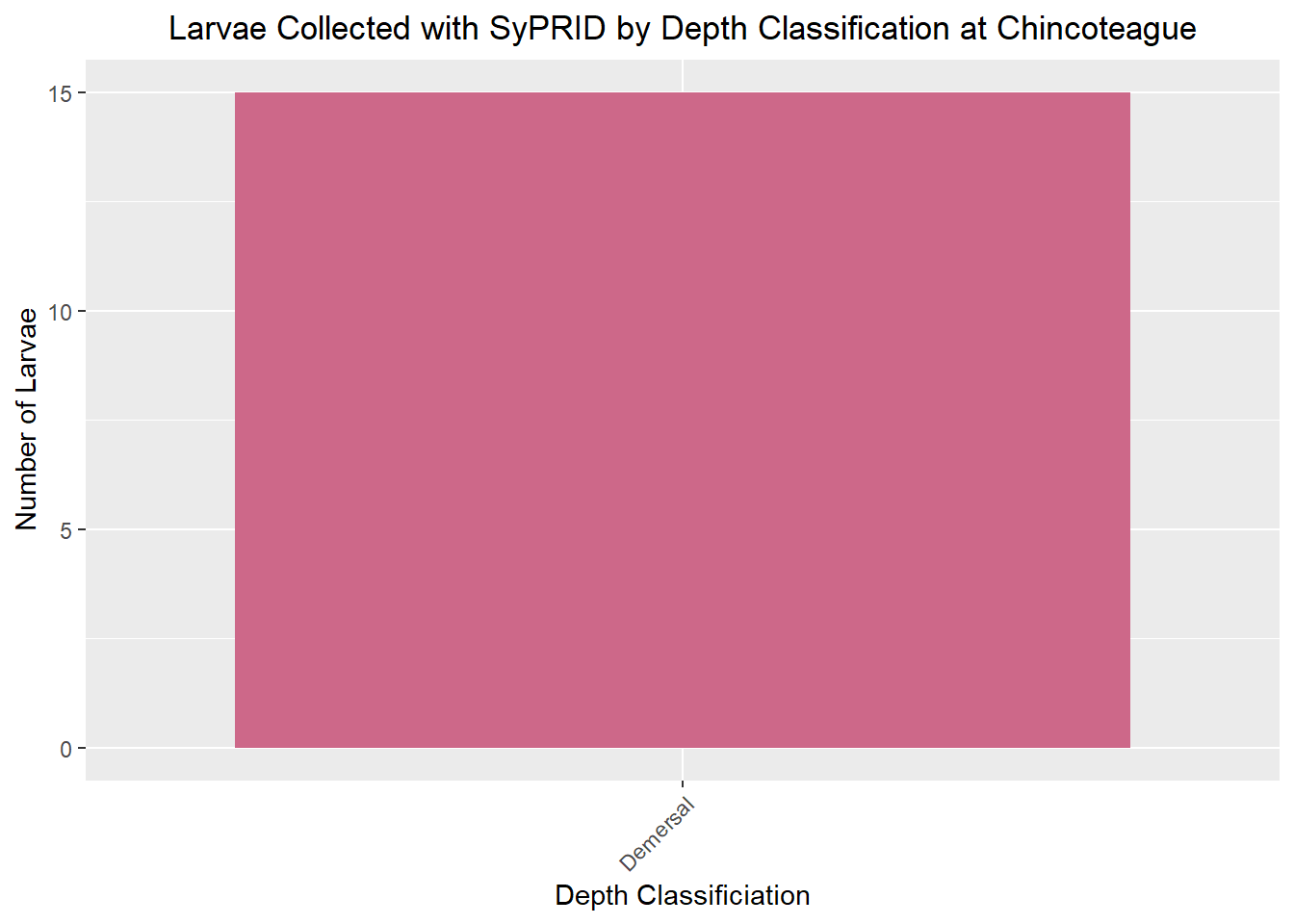
Larval Density
Den(Chincoteague, "Chincoteague")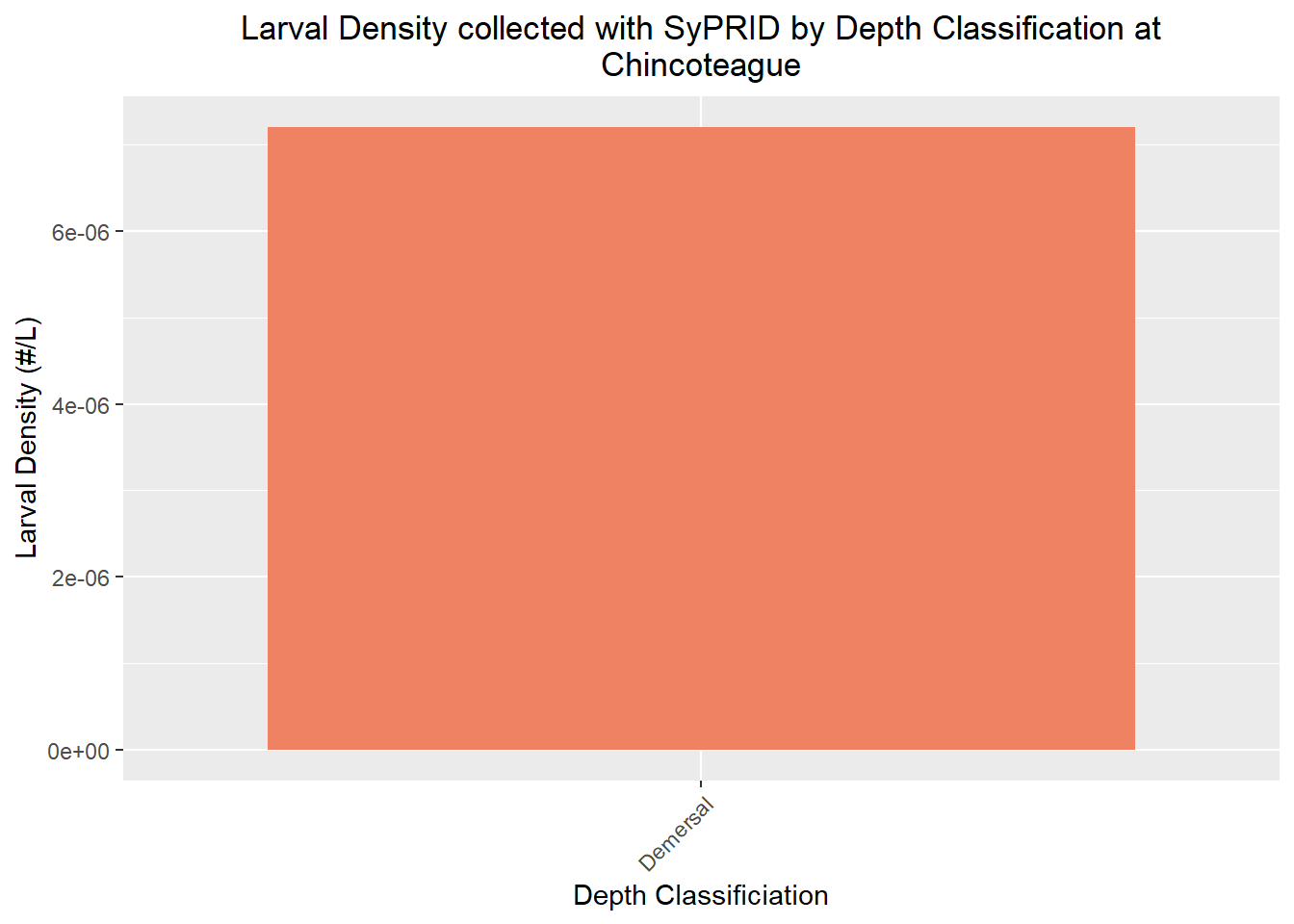
Florida Escarpment
Volume Sampled
Vol(FloridaEscarpment, "Florida Escarpment")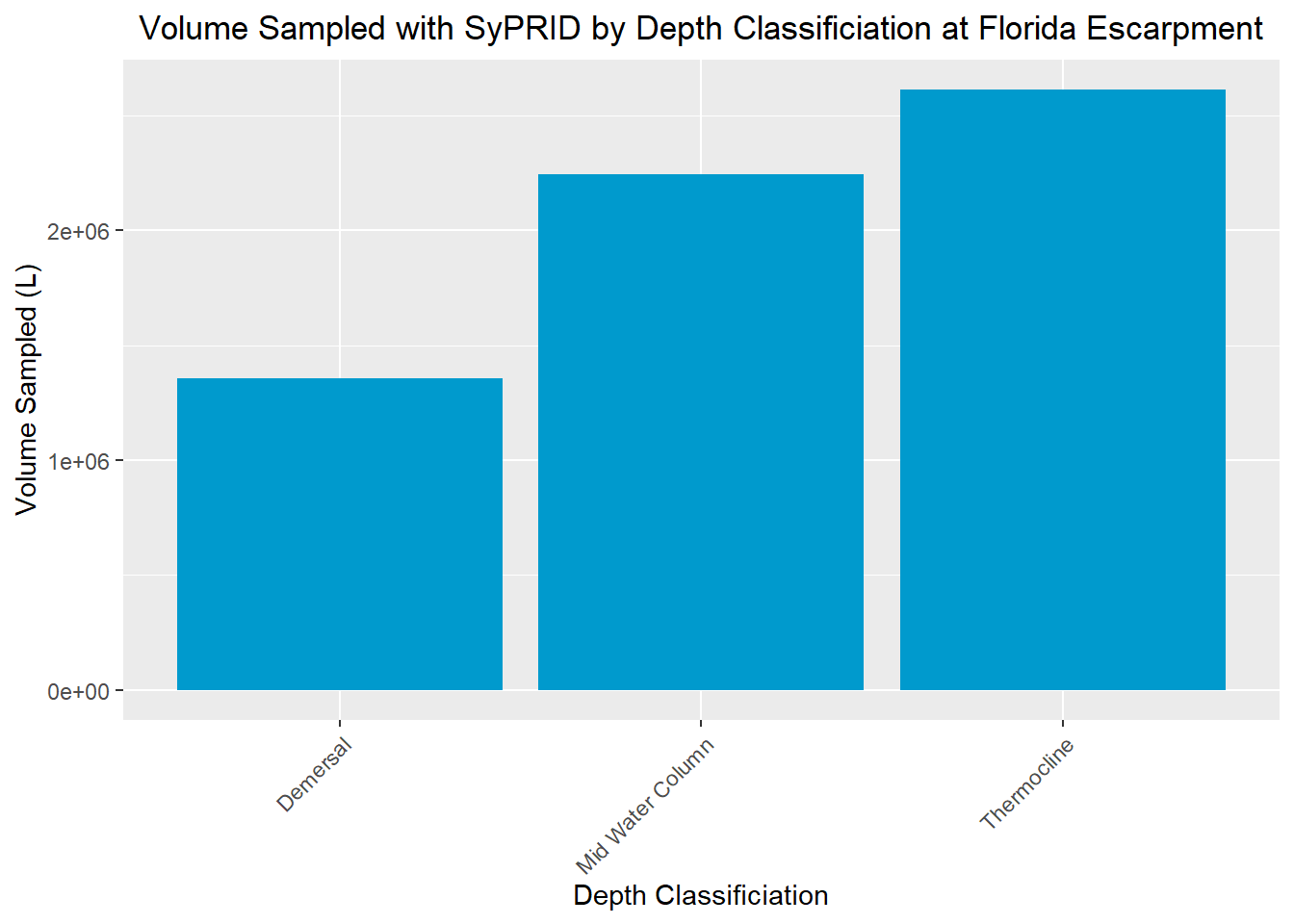
Larvae Collected
Lar(FloridaEscarpment, "Florida Escarpment")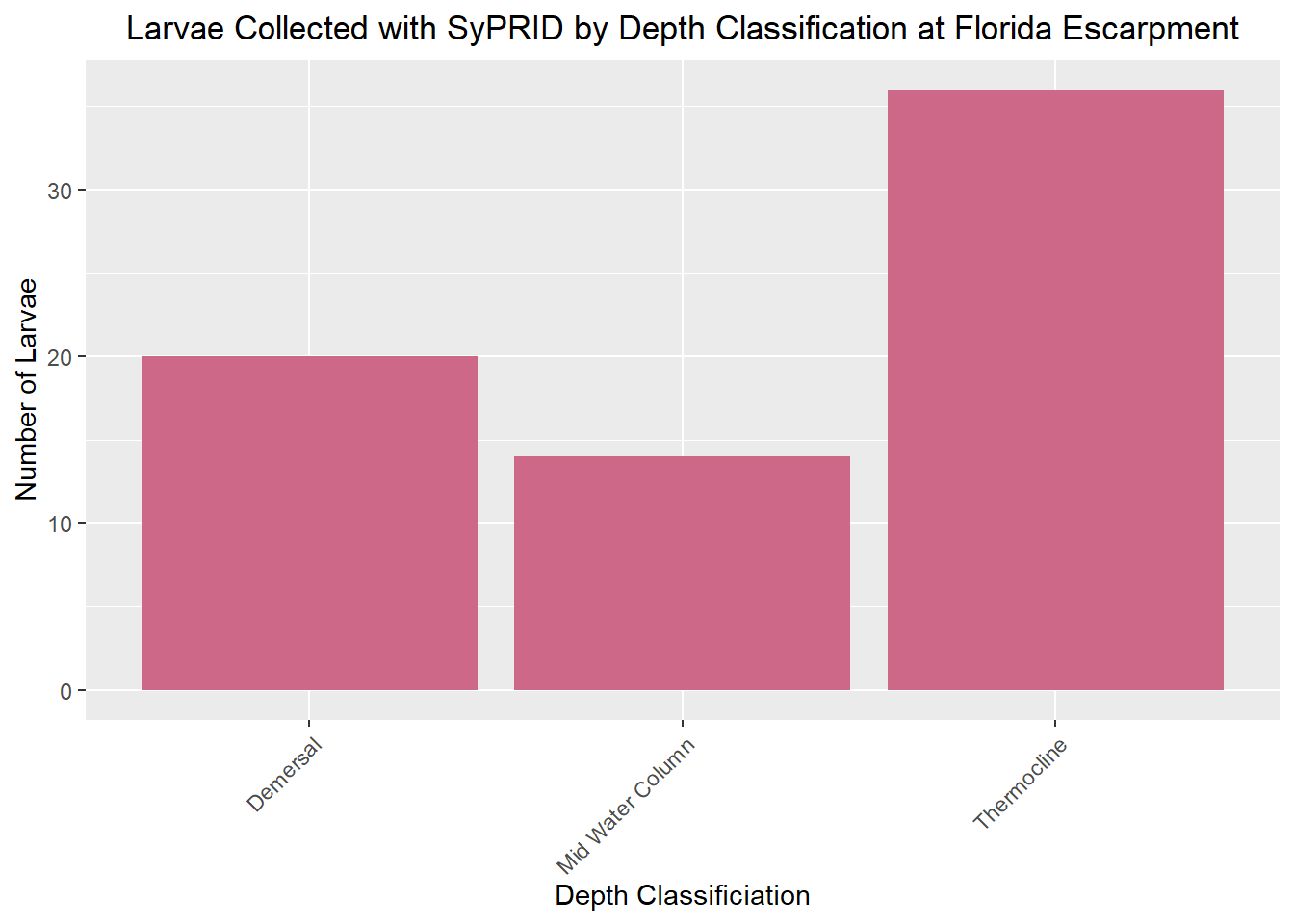
Larval Density
Den(FloridaEscarpment, "Florida Escarpment")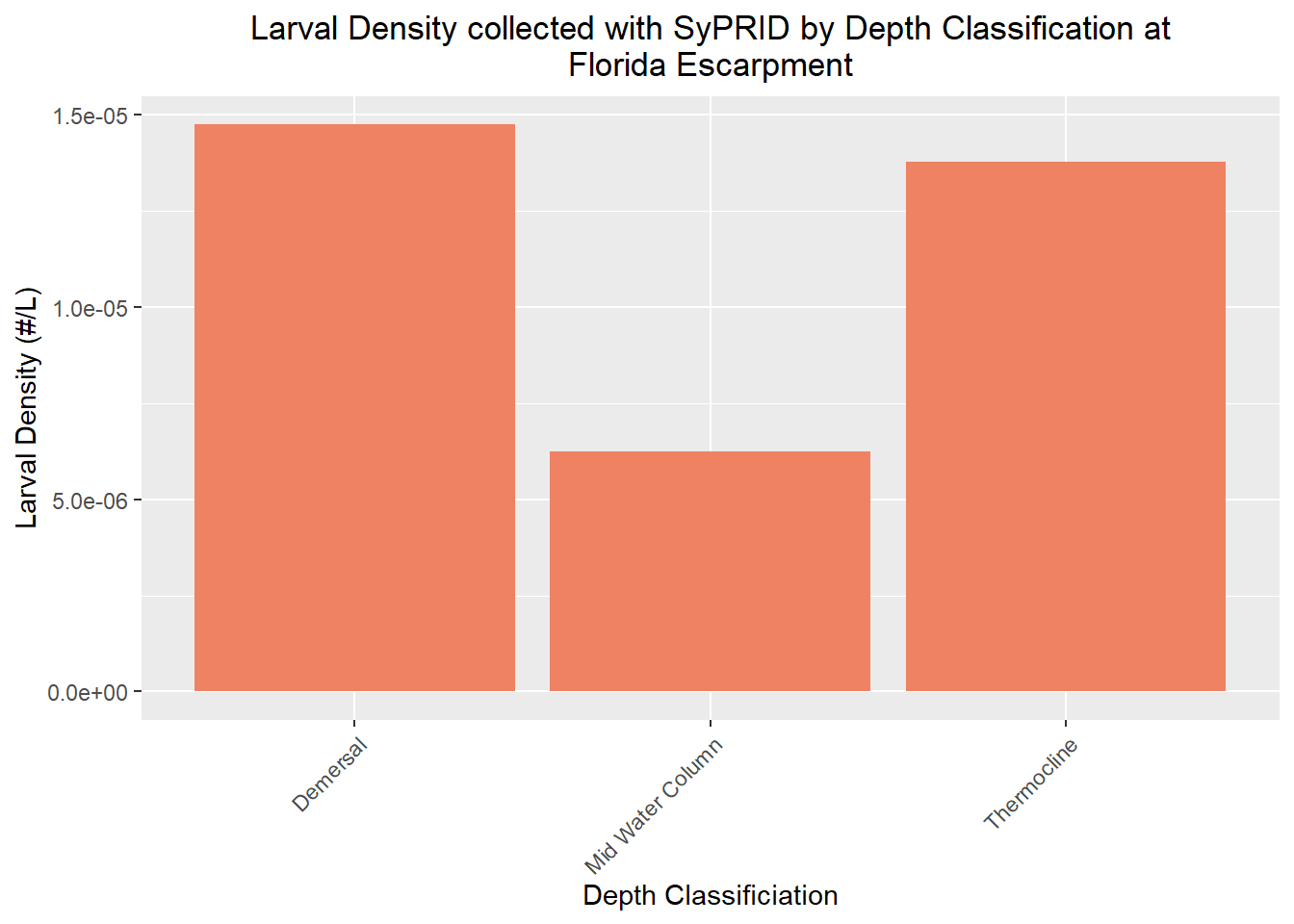
Green Canyon 234
Volume Sampled
Vol(GC234, "Green Canyon 234")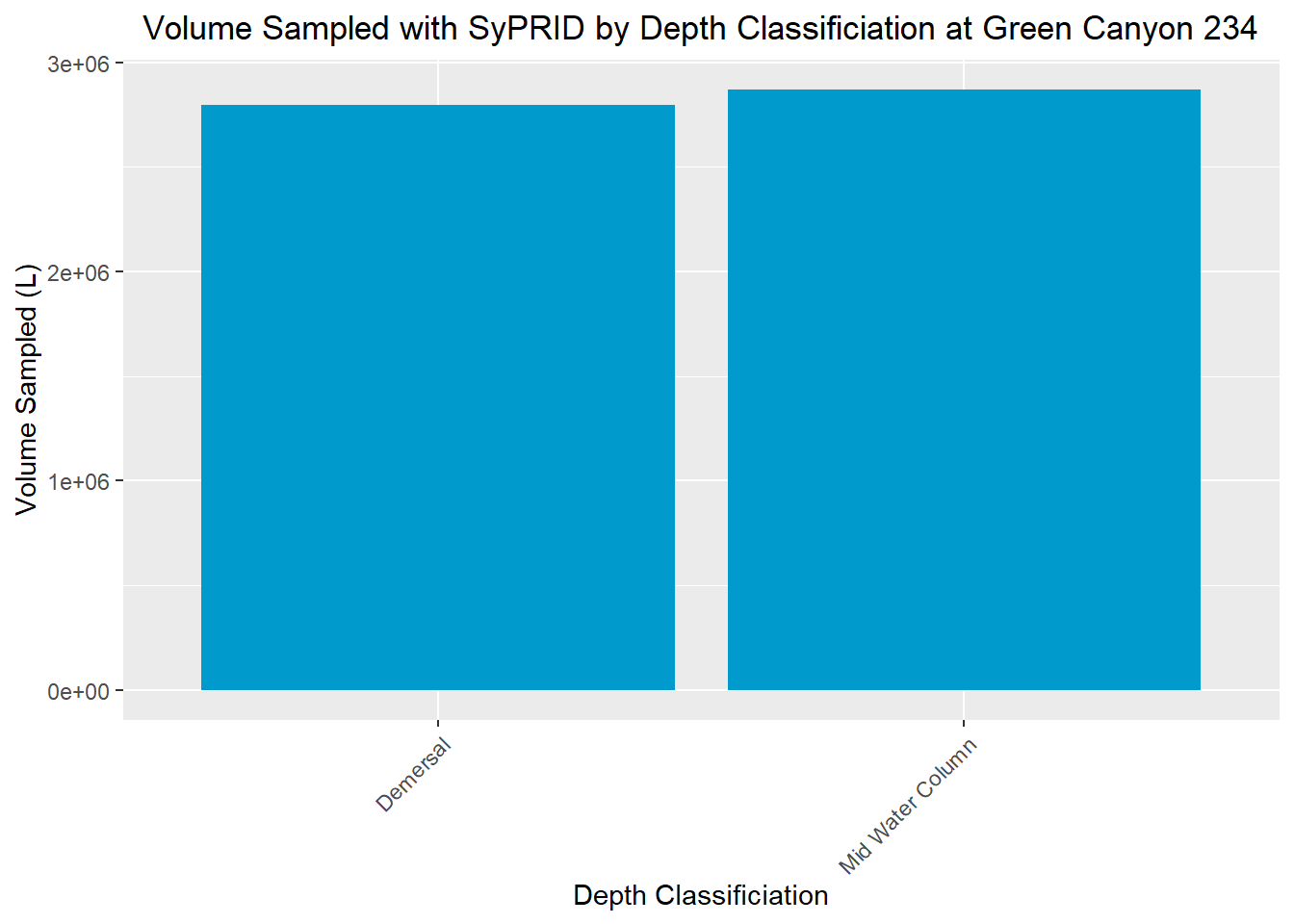
Larvae Collected
Lar(GC234, "Green Canyon 234")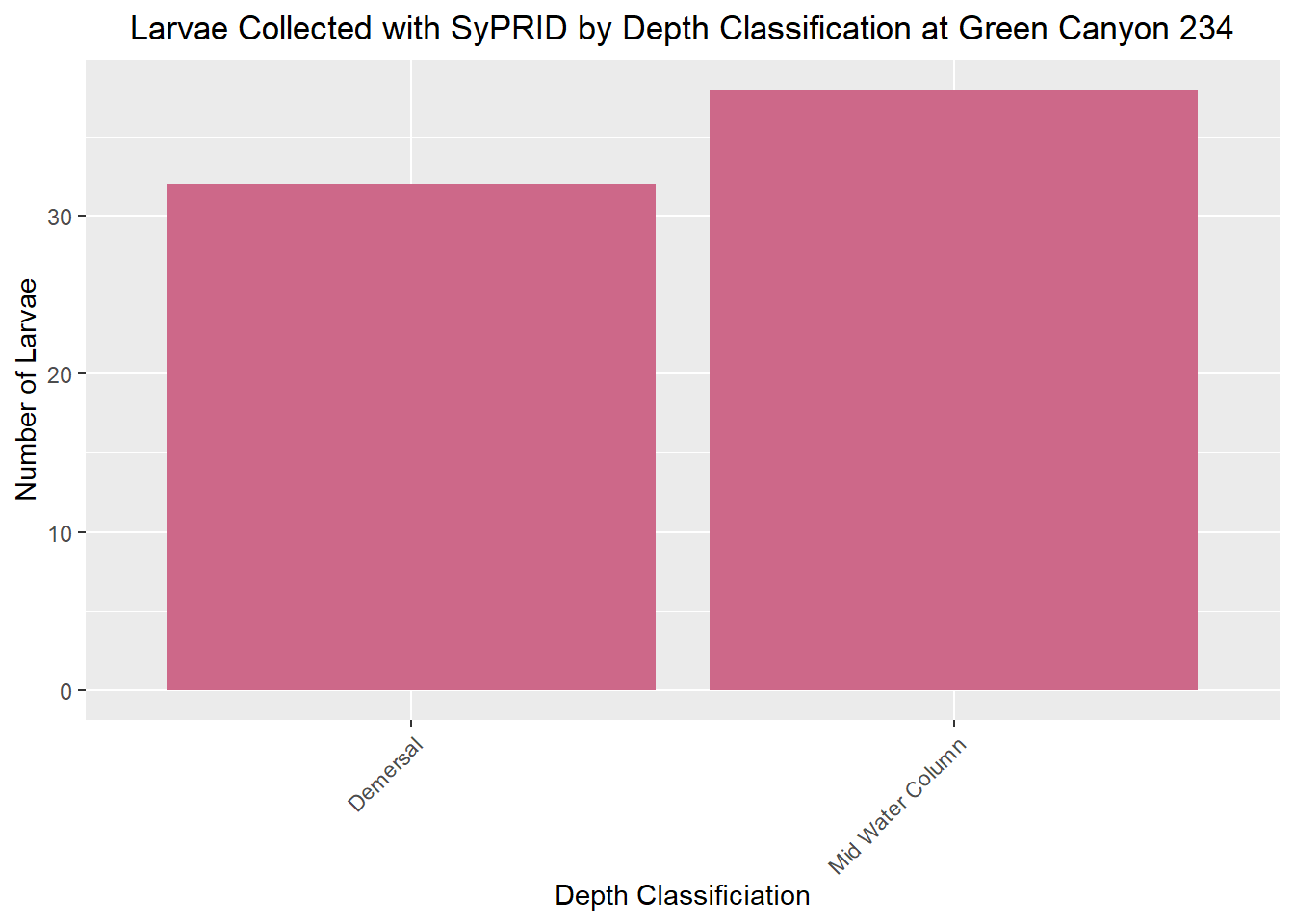
Larval Density
Den(GC234, "Green Canyon 234")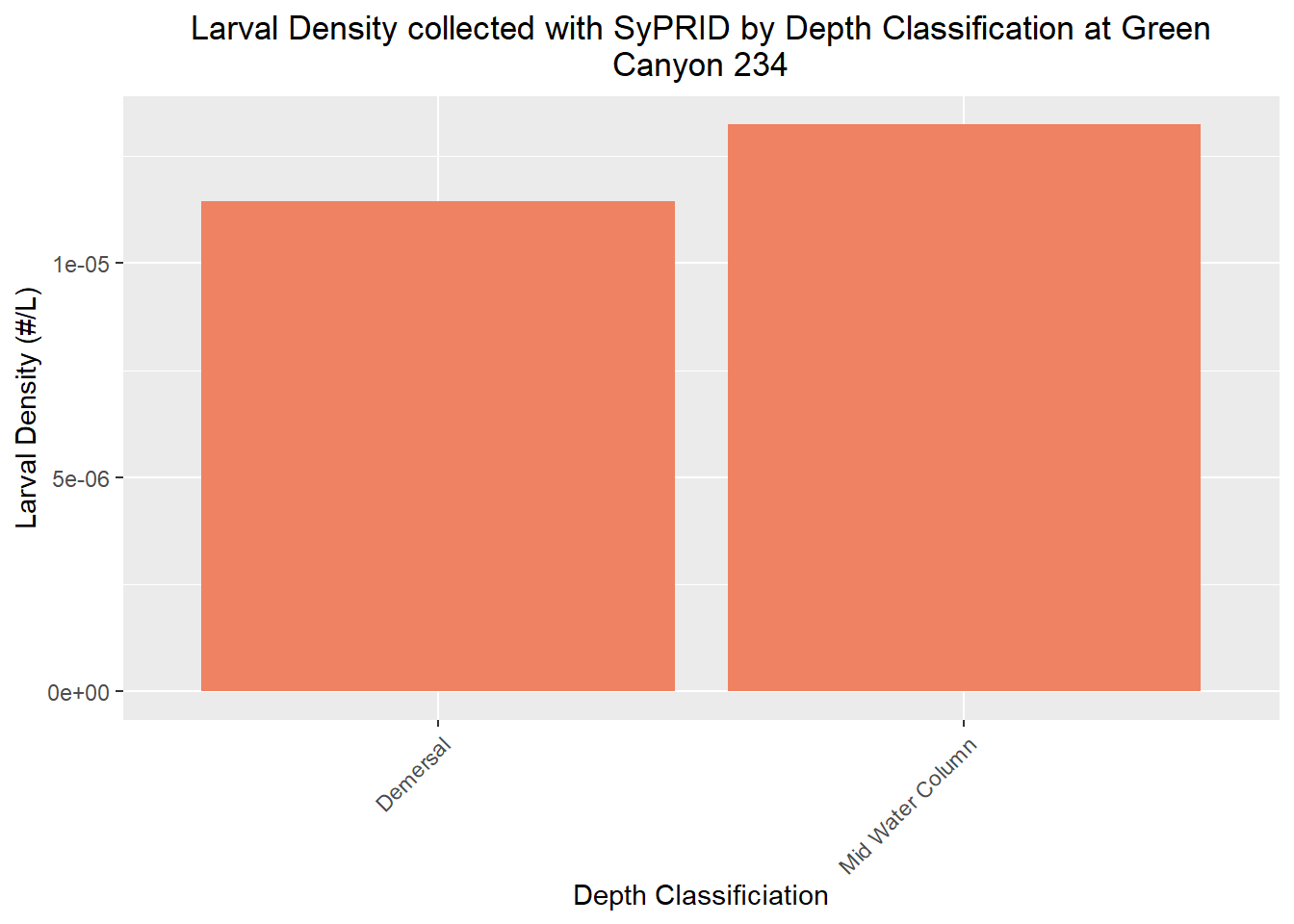
Mississippi Canyon
Volume Sampled
Vol(MississippiCanyon, "Mississippi Canyon")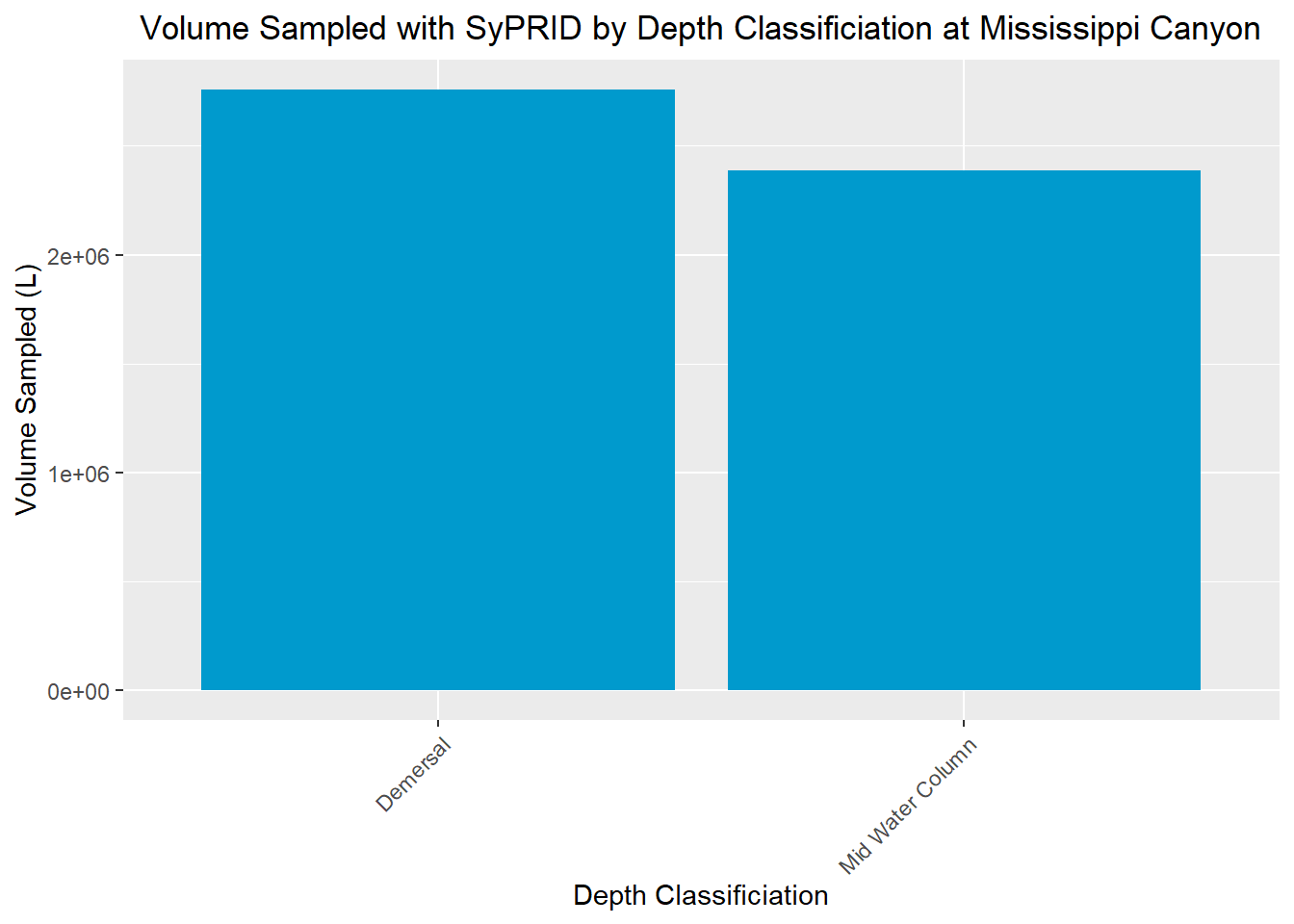
Larvae Collected
Lar(MississippiCanyon, "Mississippi Canyon")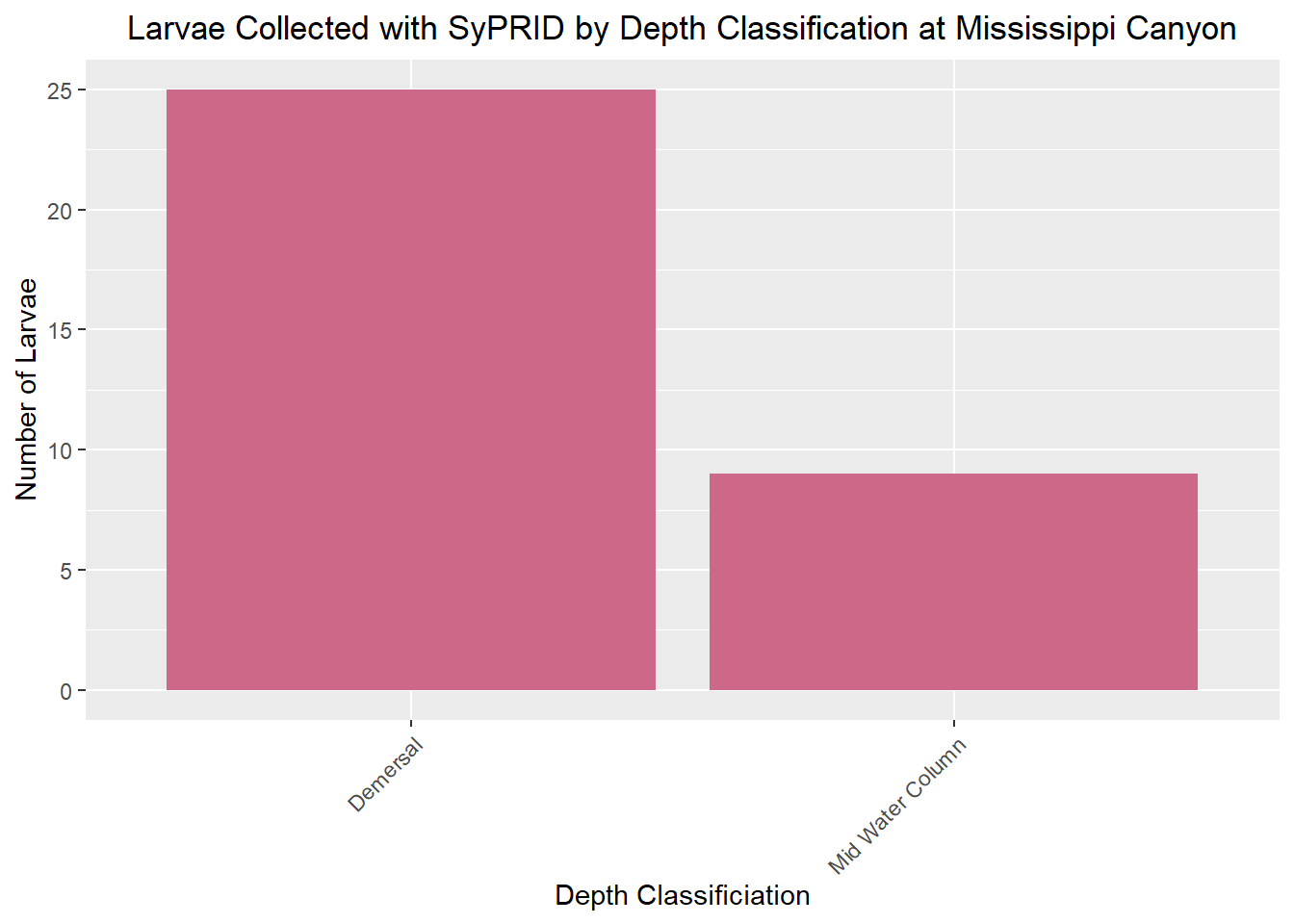
Larval Density
Den(MississippiCanyon, "Mississippi Canyon")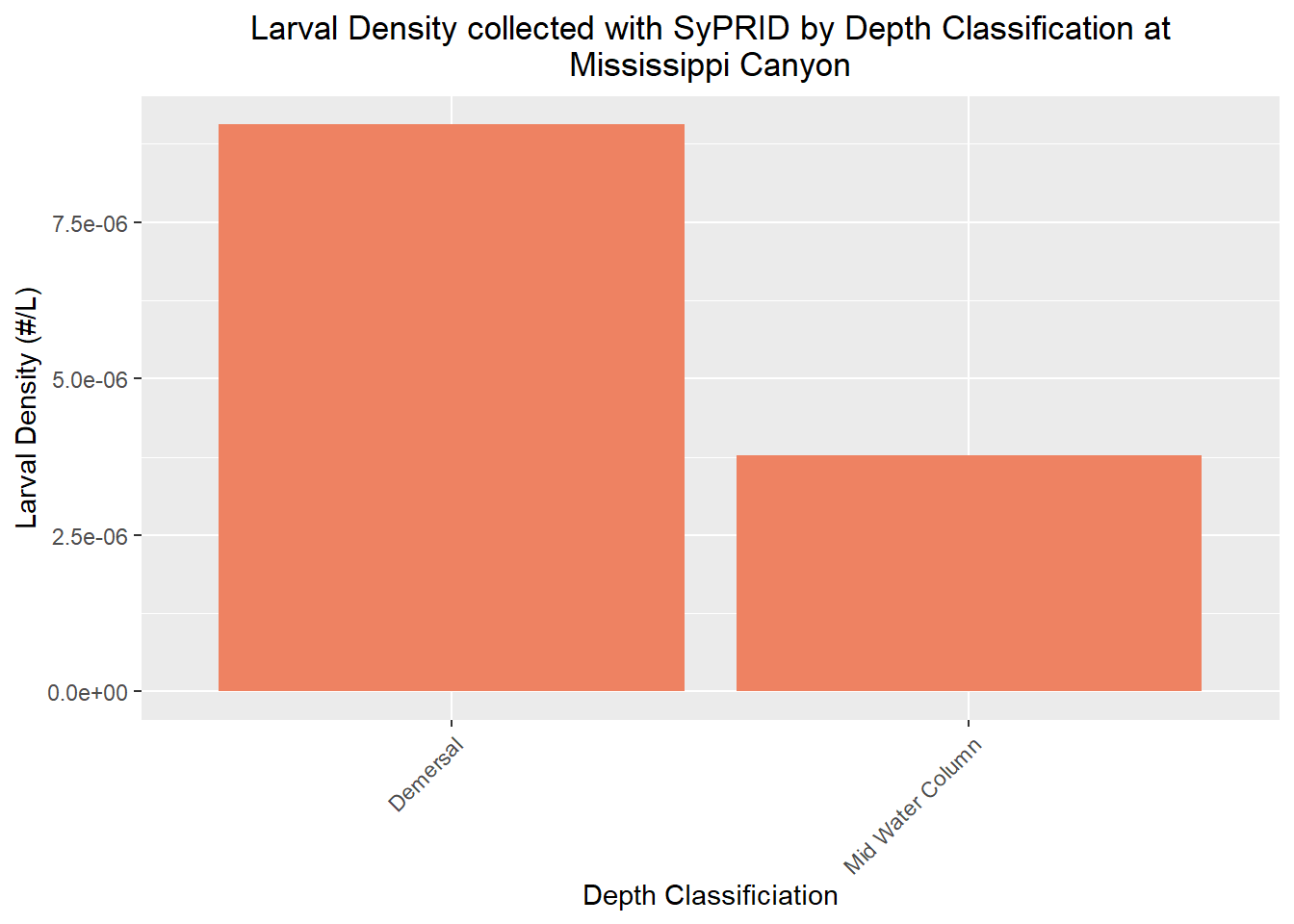
Morphotyping Results
Larval Type Guide
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
| Form | Phylum | Morphotype Name |
|---|---|---|
| AC | Phoronida | Actinotroph |
| AJ | Echinodermata | Asteroid Juvenile |
| AT | Cnidaria | Actinula |
| AU | Echinodermata | Auricularia |
| BL | Echinodermata | Brachiolaria |
| BP | Echinodermata | Bipinnaria |
| BV | Mollusca | Bivalve Veliger |
| CB | Ctenophora | Cydippid |
| CE | Cnidaria | Cerinula |
| CP | Arthropoda | Cyprid |
| CY | Bryozoa | Cyphonautes |
| DL | Echinodermata | Doliolaria |
| EC | Annelida | Echiura |
| EP | Cnidaria | Ephyra |
| GV | Mollusca | Gastropod Veliger |
| LG | Brachiopoda | Lingula |
| MI | Annelida | Mitraria |
| ML | Arthropoda | Megalopa |
| NE | Annelida | Nectochaete |
| NP | Arthropoda | Nauplius |
| OJ | Echinodermata | Ophiuroid Juvenile |
| OP | Echinodermata | Ophiopluteus |
| PC | Porifera | Parenchymula |
| PG/PS | Annelida | Pelagosphera |
| PI | Nemertea | Pilidium |
| PL | Cnidaria | Planula |
| PU | Echinodermata | Pluteus |
| SQ | Mollusca | Squid Paralarva |
| ST | Annelida | Setiger |
| TO | Hemichordata | Tornaria |
| TP | Annelida / Mollusca | Trocophore |
| UJ | Echinodermata | Urchin Juvenile |
| ZE | Cnidaria | Zoanthella |
| ZO | Arthropoda | Zoea |
Larval Type Presence by Depth
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
Demersal
| Site | Dive | # Morphotypes | Larval Types Present |
|---|---|---|---|
| Baltimore Canyon | S583P | 8 | BV/GV/NP/PC/PG/TP/ZO |
| Baltimore Canyon | S584P | 8 | BV/GV/PC/PG/PL/TP/ZO |
| Baltimore Canyon | S585P | 8 | BV/CY/PC/PL/TP/ZO |
| Blake Ridge | S587P | 7 | BV/CY/GV/MI/NE/TP |
| Blake Ridge | S588P | 7 | BV/GV/NE/OP |
| Brine Pool | S572S | 12 | BP/BV/GV/NE/NP/OJ/TP |
| Brine Pool | S592P | 13 | BV/GV/NE/OJ/PL |
| Brine Pool | S593P | 11 | AT/BL/BV/CY/GV/NE |
| Bush Hill | S594P | 11 | AT/BL/BV/GV/LG/NE/OJ/TP |
| Chincoteague | S592P | 5 | DL/NE/NP/TP/ZO |
| Florida Escarpment | S589P | 9 | BV/CY/NE/NP |
| Green Canyon 234 | S591P | 12 | BV/CY/GV/ML/NE/ZO |
| Mississippi Canyon | S595P | 7 | BV/DL/GV/LG/TP |
Mid Water Column
| Site | Dive | # Morphotypes | Larval Types Present |
|---|---|---|---|
| Baltimore Canyon | S584S | 5 | CY/GV/NE/PL/TP |
| Brine Pool | S592S | 13 | AT/BV/CY/GV/NE/OK/PC/PL/TP |
| Bush Hill | S594S | 11 | AT/BV/CY/GV/MI/NE/OJ/OP/PL |
| Florida Escarpment | S590S | 5 | BV/CY/GV/NE |
| Green Canyon 234 | S591S | 11 | BL/BV/CY/GV/LG/NE/TP |
| Mississippi Canyon | S595S | 4 | AT/GV/NE/TP |
Thermocline
| Site | Dive | # Morphotypes | Larval Types Present |
|---|---|---|---|
| Baltimore Canyon | S583S | 9 | BV/GV/ML/PC/PG/PL/TP/ZO |
| Blake Ridge | S587S | 7 | AT/BV/CY/GV/PG/TP |
| Blake Ridge | S588S | 8 | BV/CY/MI/PG/PI/PL |
| Brine Pool | S593S | 9 | BL/BV/CY/DL/GV/NE/OJ |
| Florida Escarpment | S589S | 9 | BV/GV/MI/NE/PG/TP |
Larval Types Present in Every Depth Classification*:
*Across sites
- Brachiolaria (BL)
- Bivalve Veliger (BV)
- Gastropod Veliger (GV)
- Mitraria (MI)
- Nectochaete (NE)
- Parenchymula (PC)
- Planula (PL)
- Trocophore (TP)
Community Composition Analysis
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
NMDS Plot
SALT <- read_xlsx("SALT_Larval_Consensus_Final-COPY.xlsx", sheet = "Morphotype Counts by Depth")
SALT <- subset(SALT, SALT$Method == "Sentry") #Only interested in Sentry Data
SALT <- subset(SALT, SALT$Site != "Florida Keys") #Not using Florida Keys Samples
SALT <- subset(SALT, SALT$Cruise != "EN658")
#NMDS Data Analysis
NMDS <- metaMDS(SALT[c(11:151,153:154)], k=2) #Removed unkown (UNK) Morphotype## Square root transformation
## Wisconsin double standardization
## Run 0 stress 0.2082584
## Run 1 stress 0.2249806
## Run 2 stress 0.2082584
## ... New best solution
## ... Procrustes: rmse 7.123955e-06 max resid 1.933612e-05
## ... Similar to previous best
## Run 3 stress 0.2577065
## Run 4 stress 0.2082584
## ... New best solution
## ... Procrustes: rmse 9.227413e-06 max resid 2.285938e-05
## ... Similar to previous best
## Run 5 stress 0.2249805
## Run 6 stress 0.2082983
## ... Procrustes: rmse 0.01469259 max resid 0.04988133
## Run 7 stress 0.2125757
## Run 8 stress 0.2113348
## Run 9 stress 0.2082584
## ... Procrustes: rmse 6.004551e-06 max resid 1.471178e-05
## ... Similar to previous best
## Run 10 stress 0.2370726
## Run 11 stress 0.2478411
## Run 12 stress 0.2113348
## Run 13 stress 0.2466424
## Run 14 stress 0.2082983
## ... Procrustes: rmse 0.01465319 max resid 0.04974242
## Run 15 stress 0.2082983
## ... Procrustes: rmse 0.01466932 max resid 0.04980125
## Run 16 stress 0.2367192
## Run 17 stress 0.2082584
## ... Procrustes: rmse 9.743249e-06 max resid 2.097695e-05
## ... Similar to previous best
## Run 18 stress 0.2113348
## Run 19 stress 0.2459374
## Run 20 stress 0.2100642
## *** Solution reached#Creating a data frame with the NMDS values
data.scores <- as.data.frame(scores(NMDS,NMDS$sites)) #Using scores functions from Vegan to extract NMDS values into data frame
data.scores$Site <- SALT$Site #Site Information from SALT Spreadsheet
data.scores$Cruise <- SALT$Cruise # Cruise EN658 or TN391
data.scores$Depth <- SALT $`Depth Classification` # Depth Classification - Demersal, Low WC, Mid WC, Thermocline
depth <- data.scores$Depth
site <- data.scores$Site
stress <-mean(round(NMDS$stress,digits=1))
p2 <- ggplot() +
geom_point(data=data.scores,aes(x=NMDS1,y=NMDS2,shape=data.scores$Depth, color=data.scores$Site), size=4) + # add the point markers
coord_equal() + scale_color_manual(values=c("Baltimore Canyon" = "firebrick1", "Blake Ridge" = "forest green", "Brine Pool" = "blue", "Bush Hill" = "orange", "Chincoteague" = "darkgoldenrod4", "Florida Escarpment" = "plum2", "Green Canyon 234" = "purple3", "Mississippi Canyon" = "cyan4"), name = "Site") +
scale_shape_manual(values=c("Demersal" = 15, "Mid Water Column" = 17, "Thermocline" = 19), name = "Depth Classification") +
guides(fill = guide_legend(order = 1, override.aes = list(shape = NA, alpha = 1)), shape = guide_legend(order = 2)) +
theme_classic() +
theme(panel.border = element_rect(colour = "black", fill = NA),
axis.text.x = element_text(size=12, colour="black"), axis.text.y = element_text(size=12, colour="black"),
axis.title.x = element_text(size=14, colour="black"), axis.title.y = element_text(size=14, colour="black"))+annotate("text",x=-0.5,y=.7,label=paste("Stress =",stress))
p2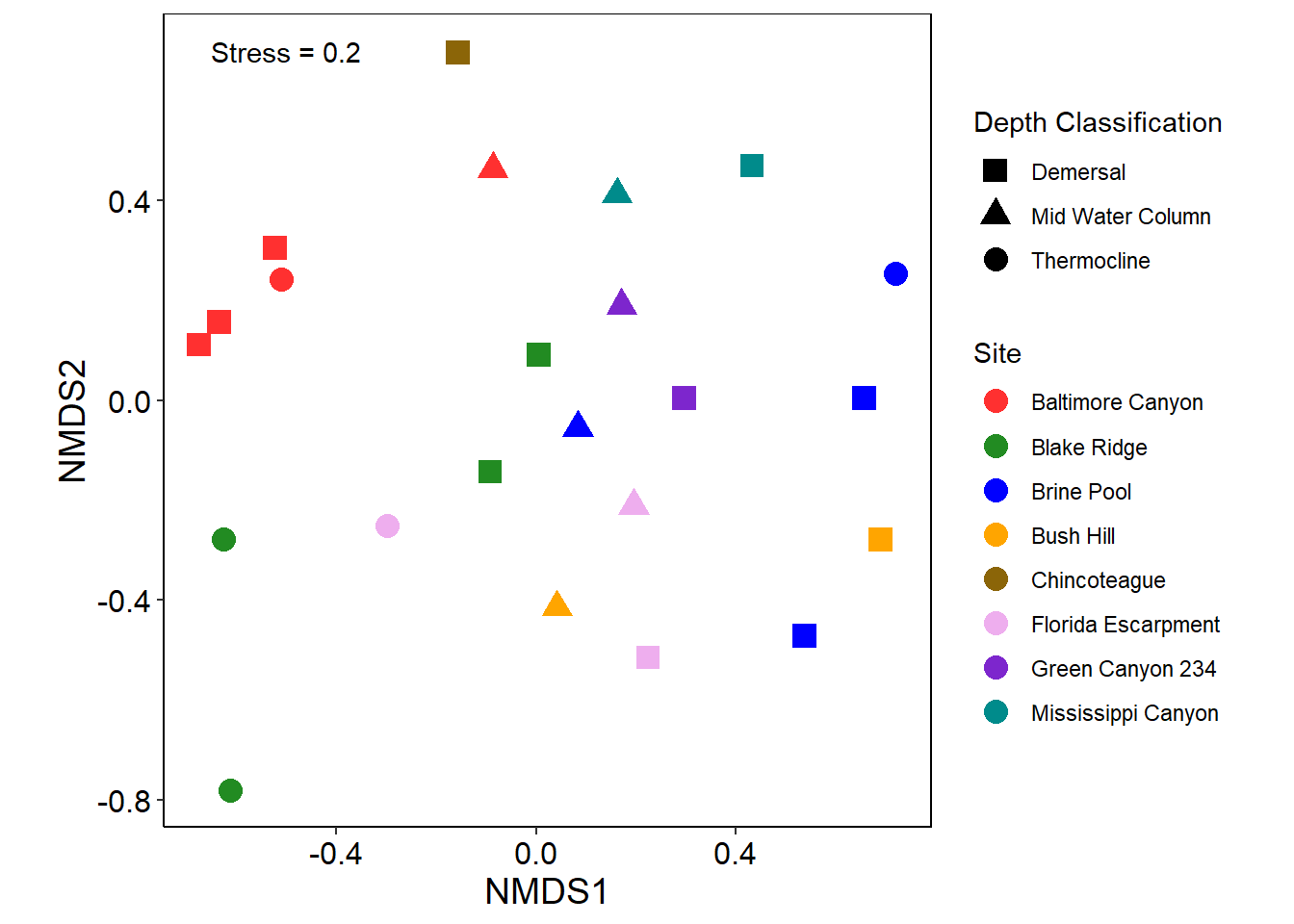
Dendogram
test.dist<-vegdist(SALT[c(11:151,153:154)], method="bray")
cluster.circles<-hclust(test.dist, method="average")
plot(cluster.circles, ylab="Dissimilarity OR 1-Similarity",labels=SALT$`Depth Classification`,hang=-.1, xlab="")
abline(h=.30,lty=3)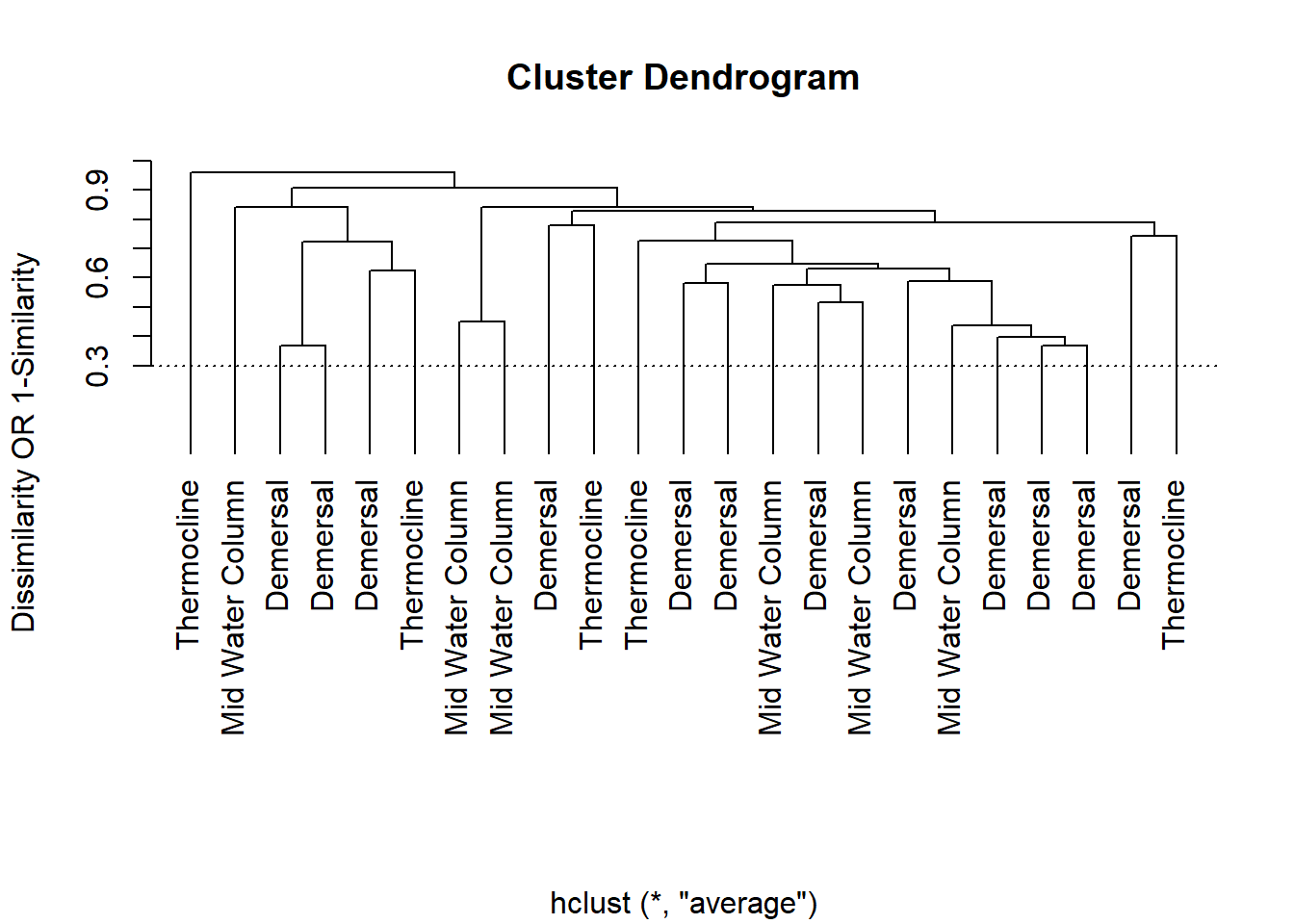
##{-}
Morphotype Proportion by Depth and Site
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
Significant Morphotypes
# Heat Map of Bivalve Veliger Morphotypes.
HeatDat <- read_xlsx("Morphotype_Consensus_Percentages.xlsx", sheet = "Final")
HeatDat <- subset(HeatDat, HeatDat$Cruise == "TN391")
HeatMat <- HeatDat[-c(1:6,35)]
mypalette <- colorRampPalette(c("white","slateblue1","slateblue4"))(n=1000)
Sig <- HeatMat[-c(1:2,6:8,11:17,19:21,23,26:28)]
rownames(Sig) <- c(HeatDat$`Sample Name`)
heat <- as.matrix(Sig)
heat <-t(heat)
heatmap.2(heat,
symkey = FALSE,
Rowv = FALSE,
Colv = FALSE,
xlab = "Site",
ylab="Morphotype",
ColSideColors = c('dodgerblue4','darkslategray3','cyan2','dodgerblue4','dodgerblue4','cyan2','dodgerblue4','darkslategray3','cyan2','dodgerblue4','darkslategray3','dodgerblue4','darkslategray3','cyan2','dodgerblue4','darkslategray3','dodgerblue4','darkslategray3'),
col = mypalette,
add.expr = c(abline(v=6.5),
col = c("cyan","darkslategray3","dodgerblue4"),pch=19),
trace="none",
density.info = "none",
key.xlab = "Proportion of Larval Sample")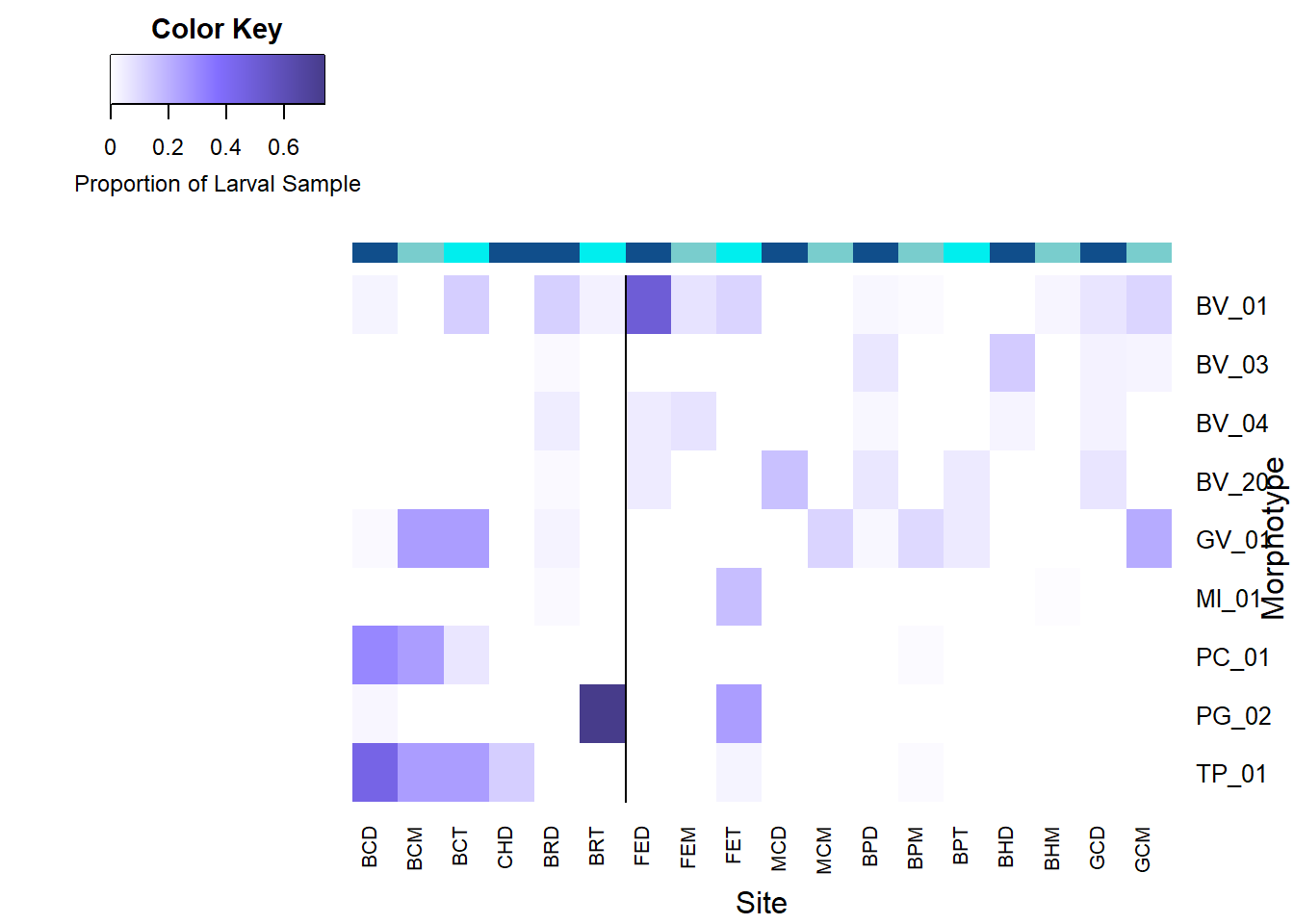
All Morphotypes
# Heat Map of Bivalve Veliger Morphotypes.
HeatDat <- read_xlsx("Morphotype_Consensus_Percentages.xlsx", sheet = "Final")
HeatDat <- subset(HeatDat, HeatDat$Cruise == "TN391")
HeatMat <- HeatDat[-c(1:6,35)]
mypalette <- colorRampPalette(c("white","slateblue1","slateblue4"))(n=1000)
row.names(HeatMat) <- (HeatDat$`Sample Name`)
heat <- as.matrix(HeatMat)
heat <-t(heat)
heatmap.2(heat,
symkey = FALSE,
Rowv = FALSE,
Colv = FALSE,
xlab = "Site",
ylab="Morphotype",
ColSideColors = c('dodgerblue4','darkslategray3','cyan2','dodgerblue4','dodgerblue4','cyan2','dodgerblue4','darkslategray3','cyan2','dodgerblue4','darkslategray3','dodgerblue4','darkslategray3','cyan2','dodgerblue4','darkslategray3','dodgerblue4','darkslategray3'),
col = mypalette,
add.expr = c(abline(v=6.5),
col = c("cyan","darkslategray3","dodgerblue4"),pch=19),
trace="none",
density.info = "none",
key.xlab = "Proportion of Larval Sample")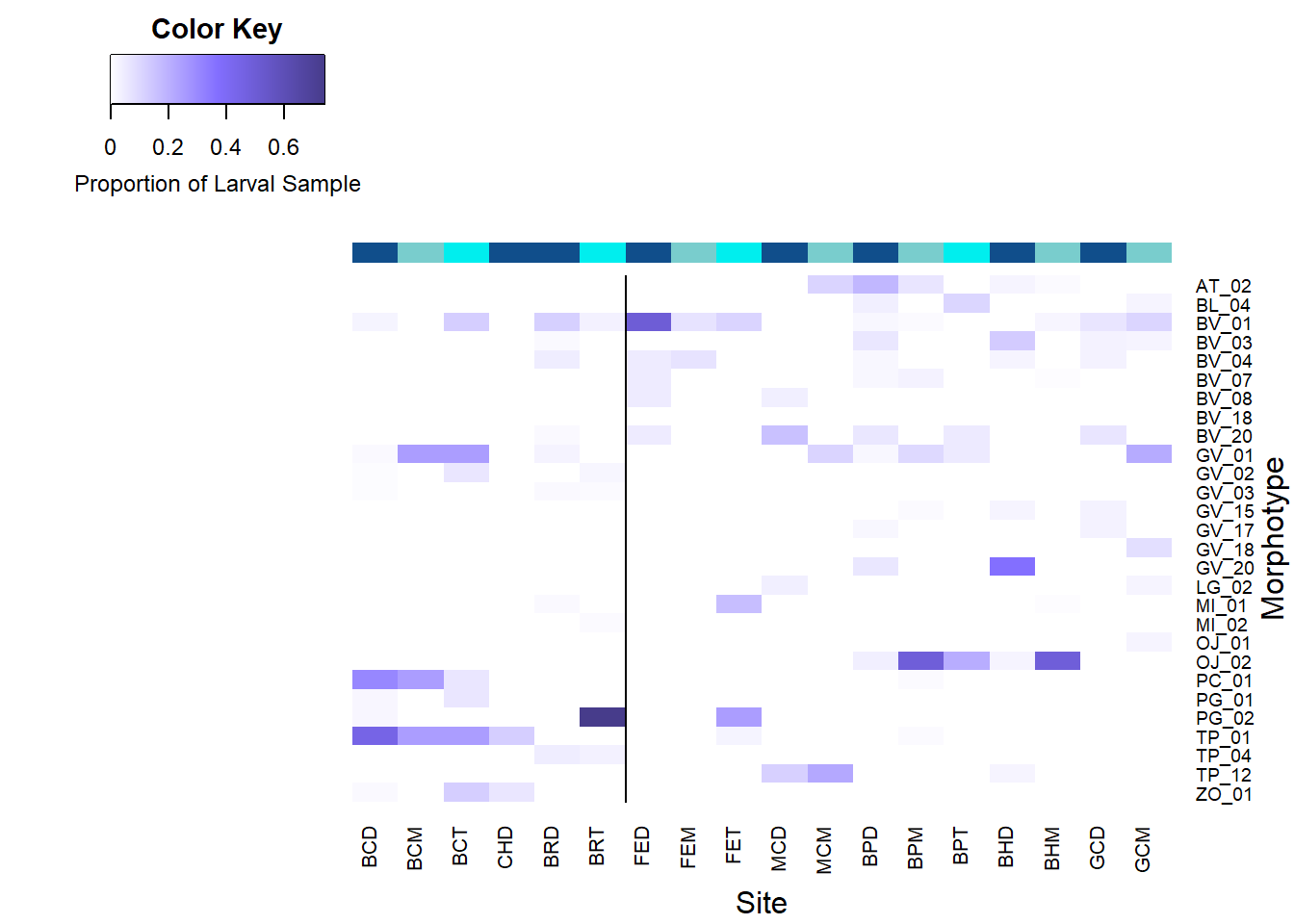
Brine Pool Seasonality
# Heat Map of Bivalve Veliger Morphotypes.
HeatDat <- read_xlsx("Morphotype_Consensus_Percentages.xlsx", sheet = "Final")
HeatDat <- subset(HeatDat, HeatDat$Site == "Brine Pool")
HeatMat <- HeatDat[-c(1:6,13,17:18,24,29,32:34,35)]
mypalette <- colorRampPalette(c("white","slateblue1","slateblue4"))(n=1000)
row.names(HeatMat) <- (c("FallD","FallM","SpringD","SpringM","SpringT"))
heat <- as.matrix(HeatMat)
heat <-t(heat)
heatmap.2(heat,
symkey = FALSE,
Rowv = FALSE,
Colv = FALSE,
xlab = "Site",
ylab="Morphotype",
ColSideColors = c('dodgerblue4','darkslategray3','dodgerblue4','darkslategray3','cyan2'),
col = mypalette,
add.expr = c(abline(v=2.5),
col = c("cyan","darkslategray3","dodgerblue4"),pch=19),
trace="none",
density.info = "none",
key.xlab = "Proportion of Larval Sample",
srtCol=45)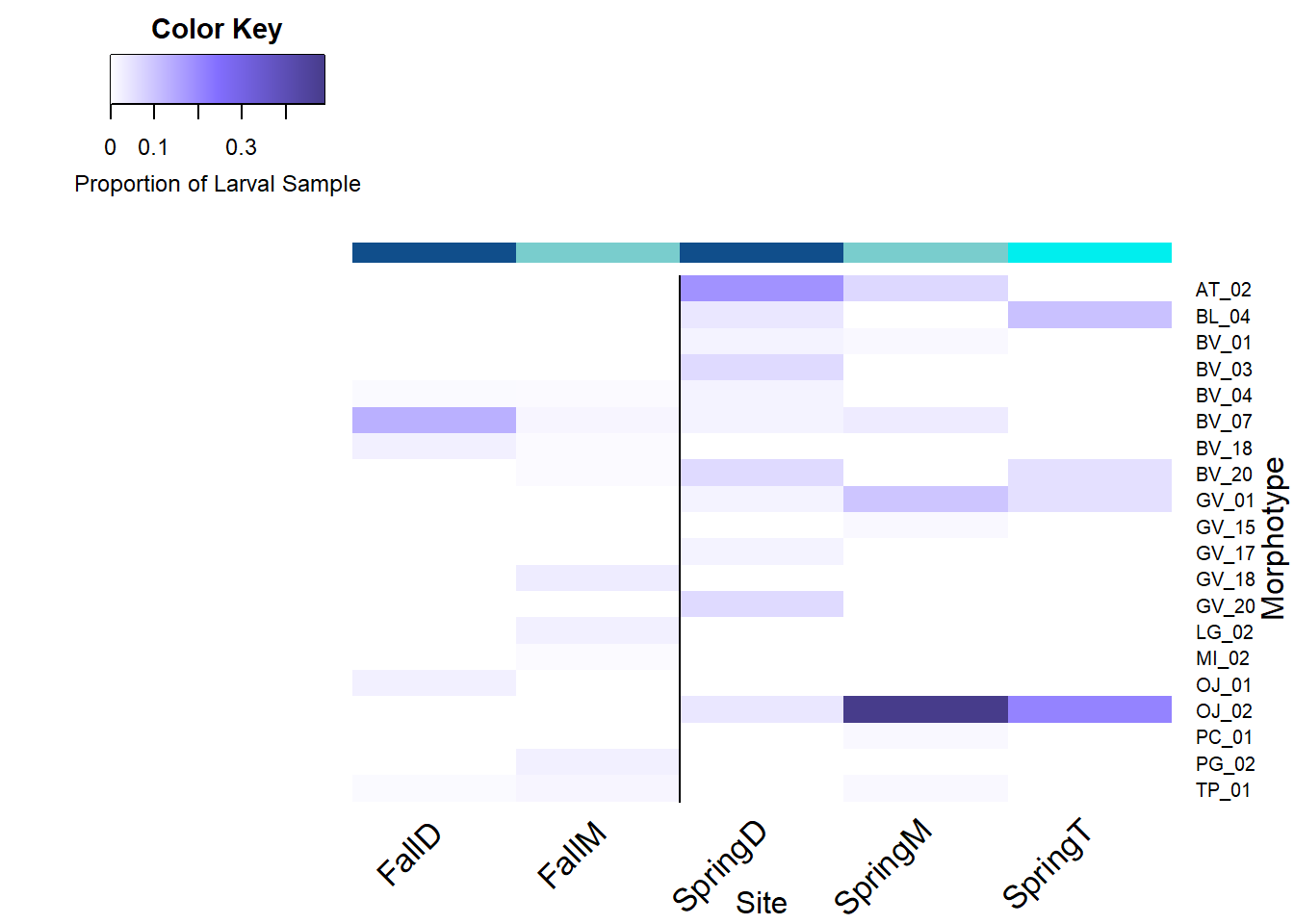
* Site codes denote the following: BC = Baltimore Canyon, CH = Chincoteague, BR = Blake Ridge, FE = Florida Escarpment, MC = Mississippi Canyon, BP = Brine Pool, BH = Bush Hill, GC = Green Canyon 234. The letters after these site codes represent the depth classificaiton; D = Demersal, M = Mid Water Column, T = Thermocline.
For Brine Pool, collections from the Fall 2020 EN658 cruise are denoted with the codes Fall, while the other site codes are from the Spring 2021 TN391 cruise.
The color bar along the columns acts as a visual aid for the depth classifications; demersal ( darkblue ), mid water column ( gray-blue ), thermocline ( cyan ).
The vertical line represents the geographical split between the Western Atlantic Margin (left) and the Gulf of Mexico (right) sites or the split between Fall and Spring cruises.
Total Counts for Depth Classifications (Notes)
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
- Many BV morphotypes have higher abundance in demersal samples
- BV02, BV03, BV04, BV07-BV12, BV14, BV15, BV20
- BV01 somewhat consistent across depths
- Gv morphotypes vary depth classifications
- Higher Demersal: GV04, GV07, GV16, GV17, GV19, GV20, GV22
- Higher Mid WC: GV09, GV10
- Higher Thermocline: GV02, GV03, GV13, GV14, GV21
- Higher Mid / Thermo: GV01
- NE most abundance in Demersal & mid water column.
- PC more abundant closer to bottom.
- PG more abundance in thermocline (except PG01 similar thermocline and demersal).
- PL more abundant higher in water column.
- TP most abundant demersally, similar low water column and thermocline.
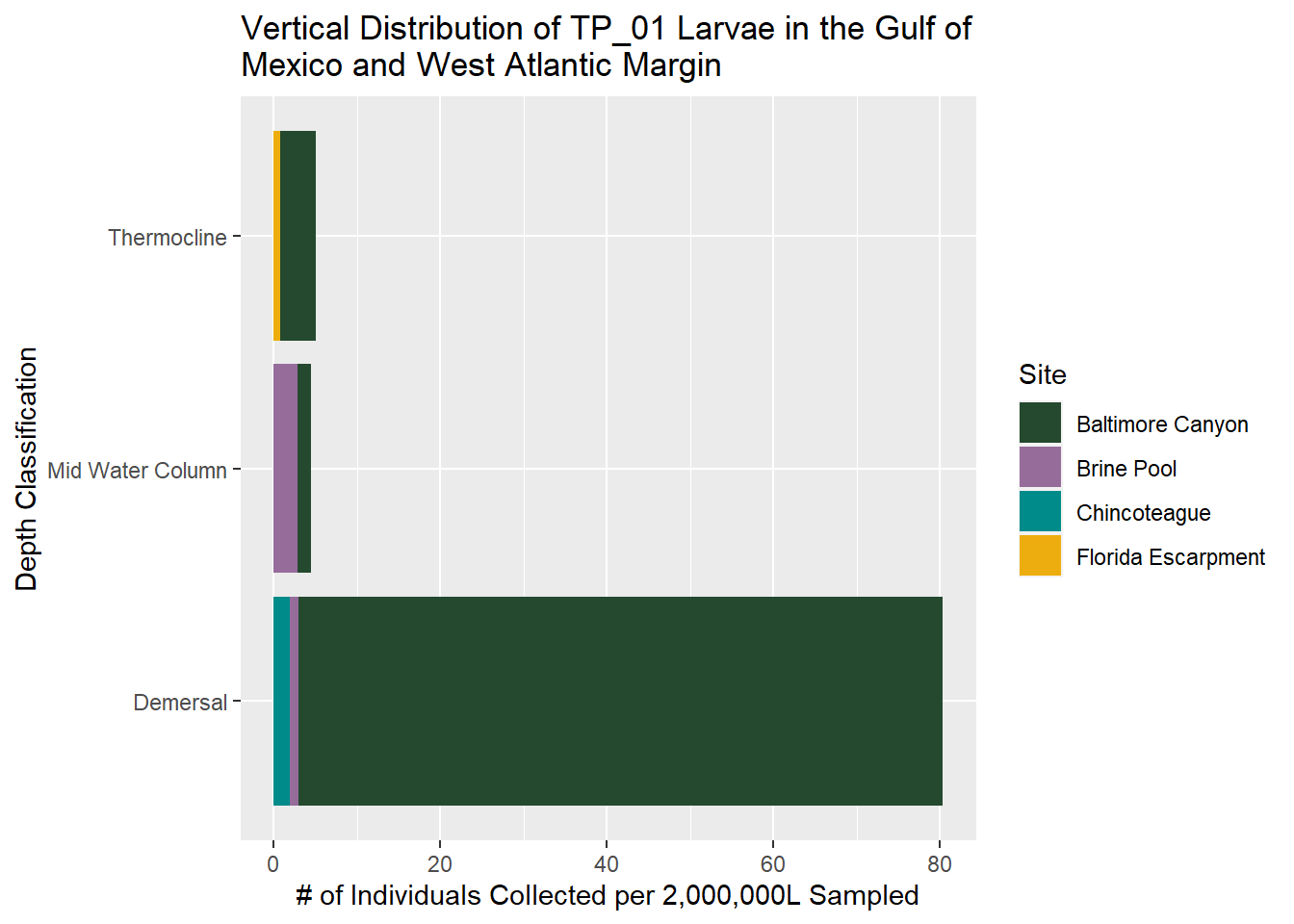
- TP01 more abundant closer to bottom.
- ZO slightly more abundant demersal to thermocline
Vertical Distributions of Target Larvae
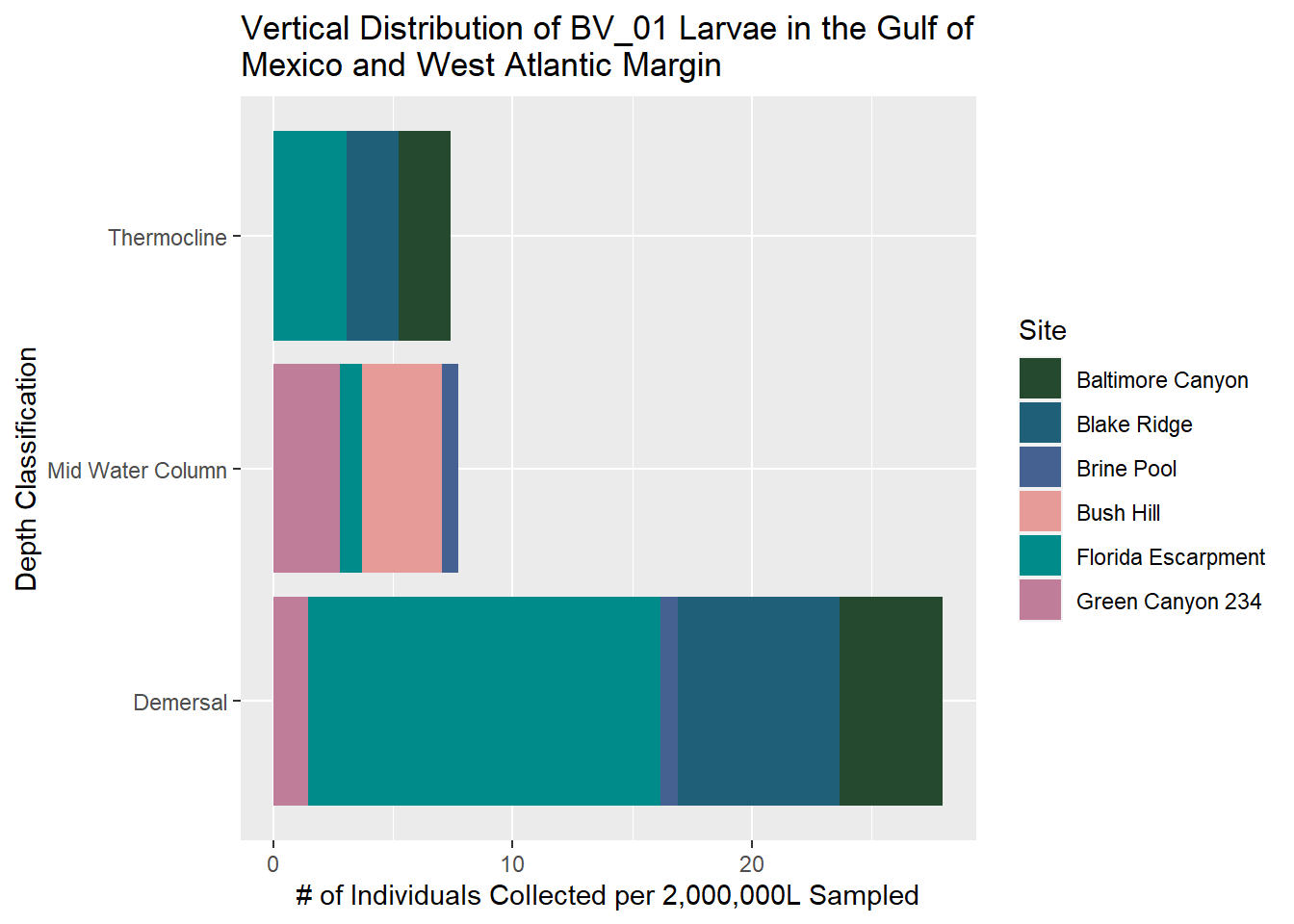
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
BV_01
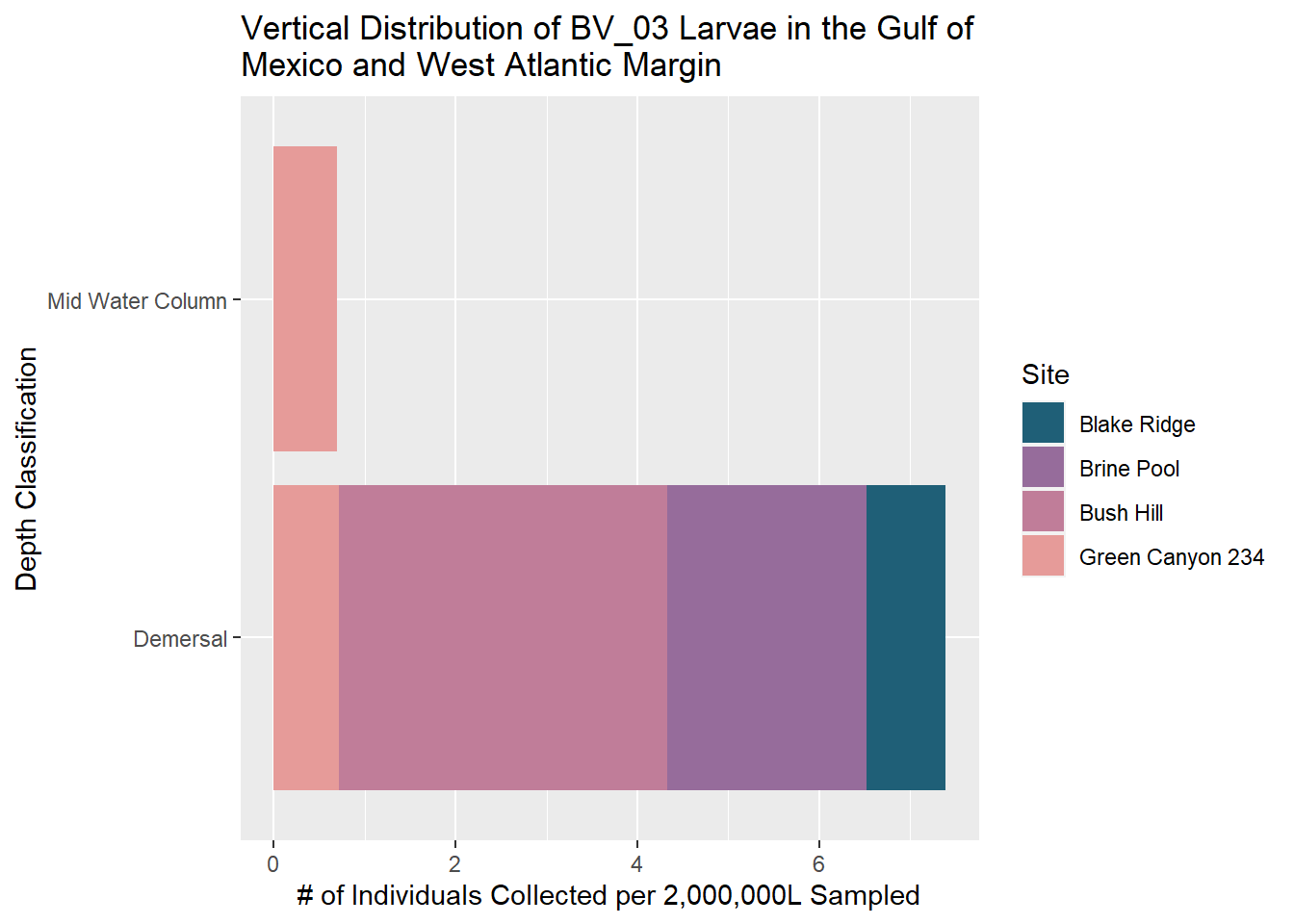
BV_03
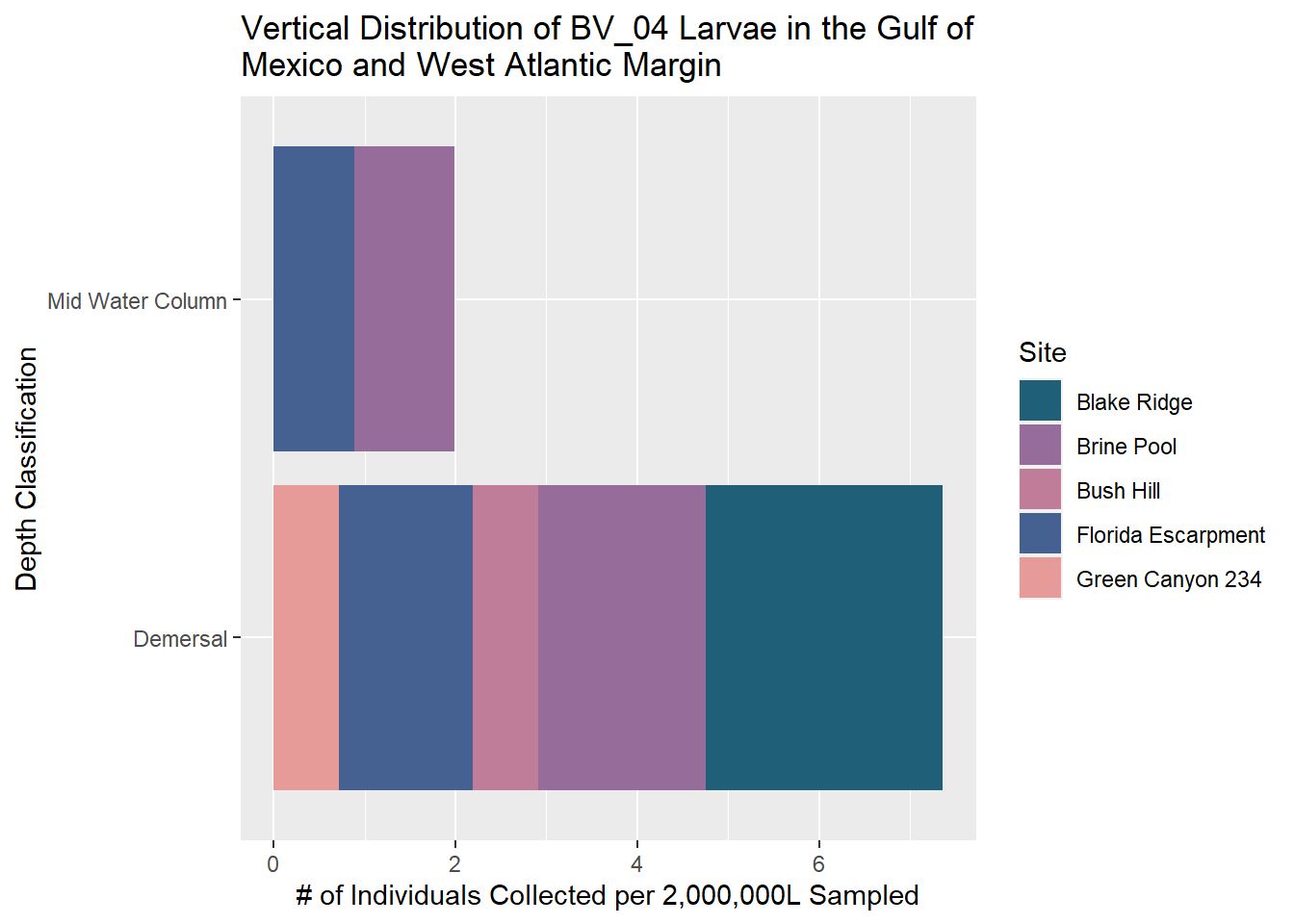
BV_04
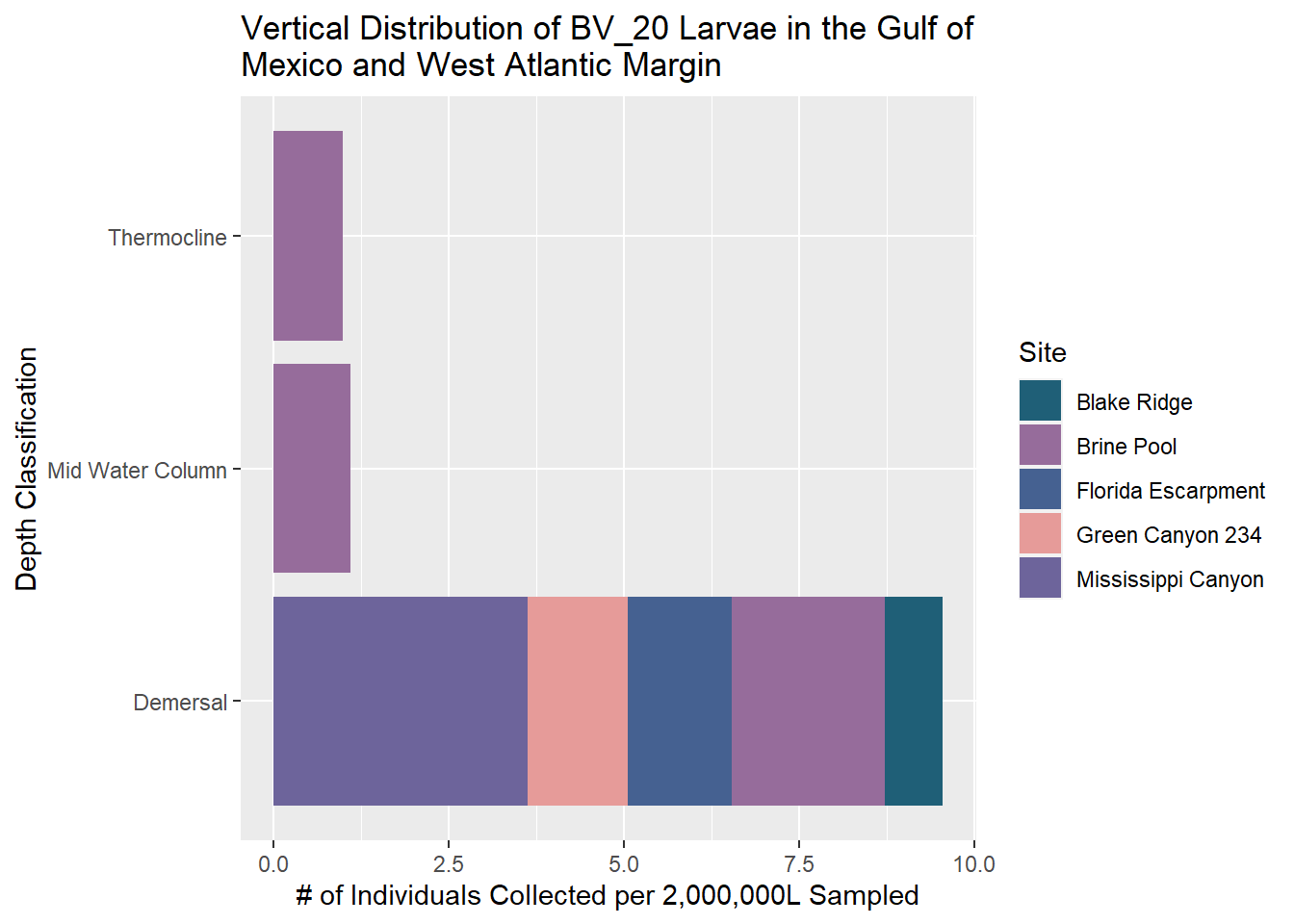
BV_20
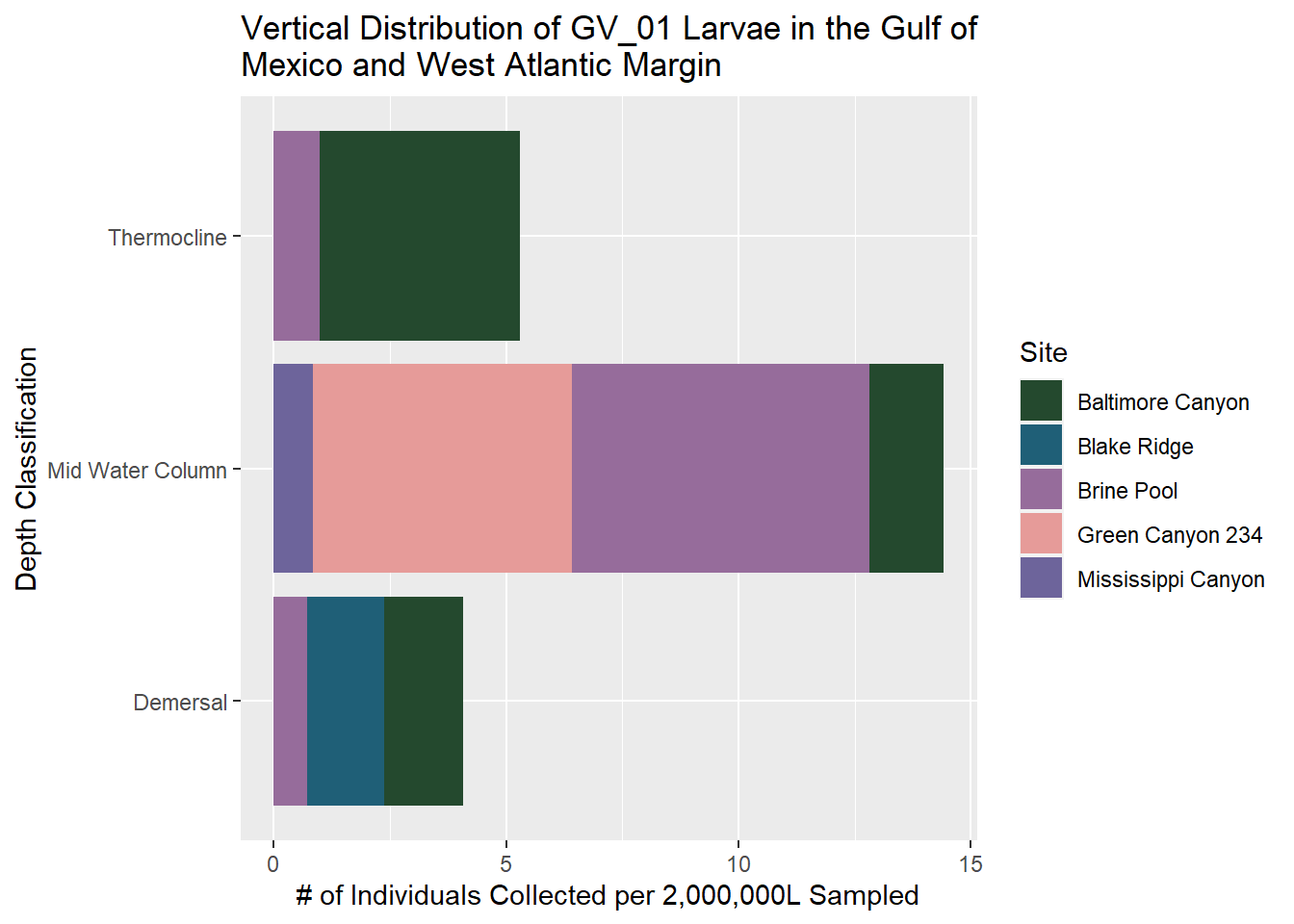
GV_01
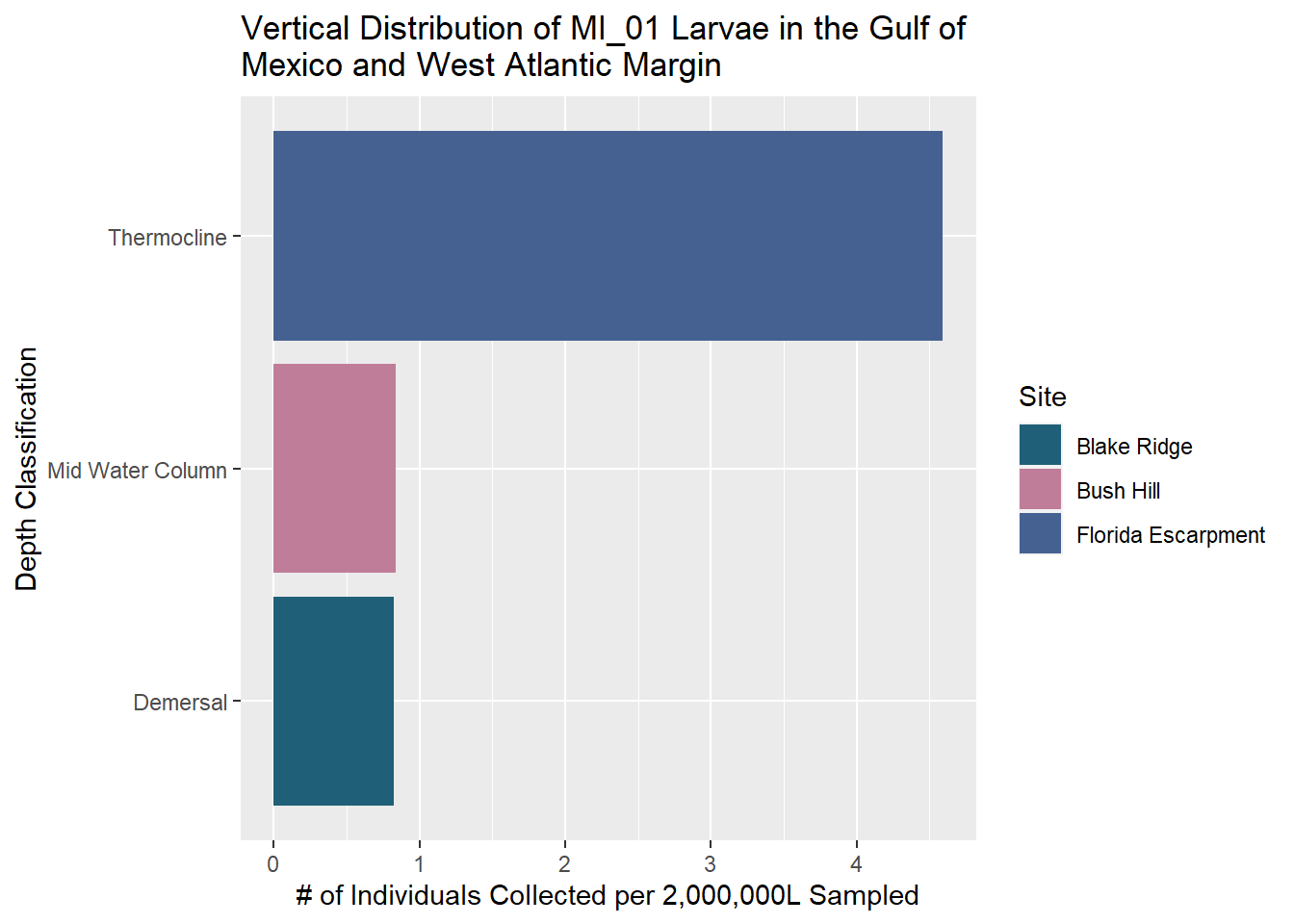
MI_01
PC_01

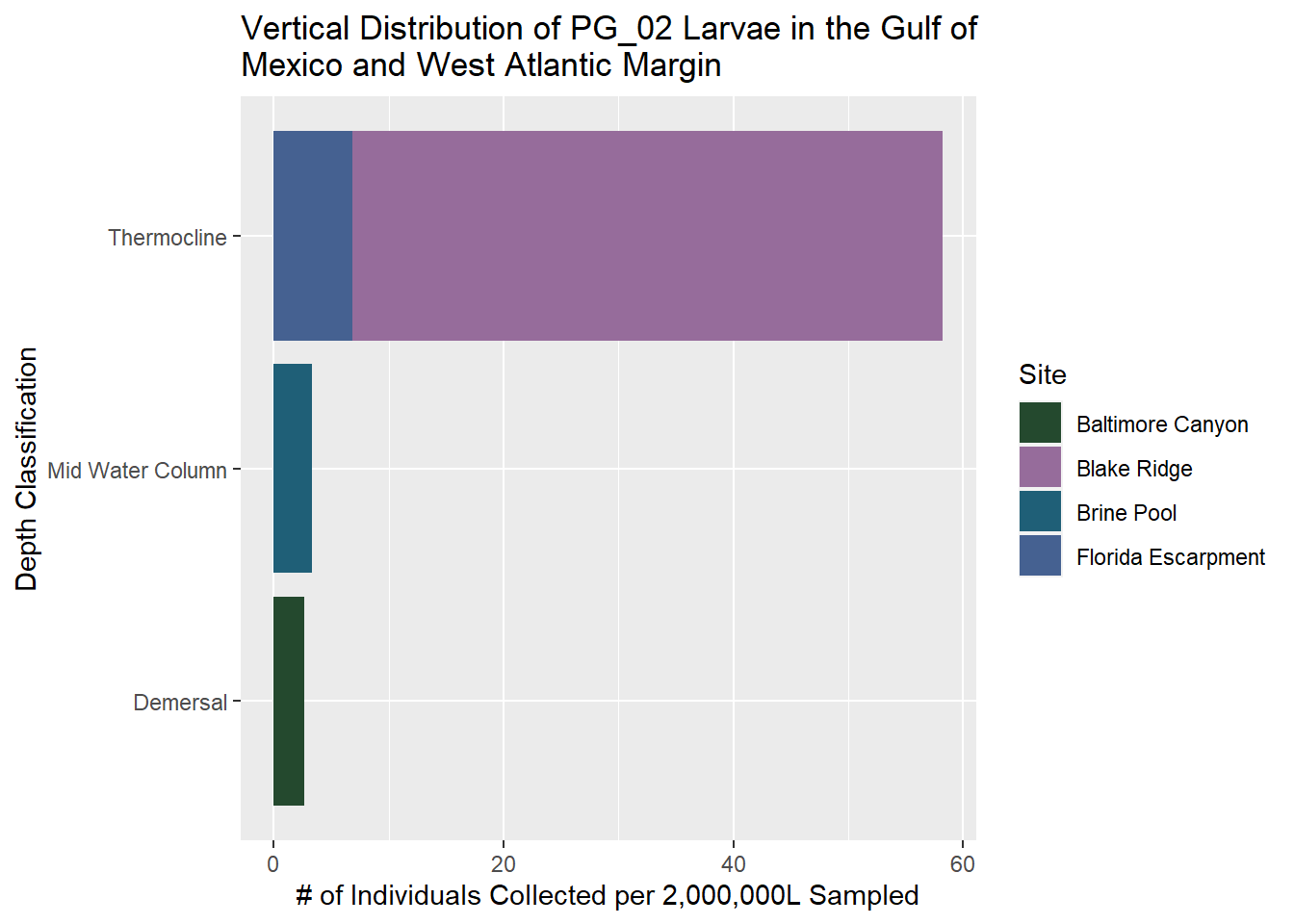
PG_02
TP_01